3WSD
 
 | | Oxidized HcgD from Methanocaldococcus jannaschii | | Descriptor: | CHLORIDE ION, FE (III) ION, Putative GTP cyclohydrolase 1 type 2 | | Authors: | Fujishiro, T, Ermler, U, Shima, S. | | Deposit date: | 2014-03-13 | | Release date: | 2014-08-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A possible iron delivery function of the dinuclear iron center of HcgD in [Fe]-hydrogenase cofactor biosynthesis
Febs Lett., 588, 2014
|
|
3WV9
 
 | | Guanylylpyridinol (GP)- and ATP-bound HcgE from Methanothermobacter marburgensis | | Descriptor: | 5'-O-[(S)-{[2-(carboxymethyl)-6-hydroxy-3,5-dimethylpyridin-4-yl]oxy}(hydroxy)phosphoryl]guanosine, ADENOSINE-5'-TRIPHOSPHATE, Hmd co-occurring protein HcgE, ... | | Authors: | Fujishiro, T, Ermler, U, Shima, S. | | Deposit date: | 2014-05-16 | | Release date: | 2015-04-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Protein-pyridinol thioester precursor for biosynthesis of the organometallic acyl-iron ligand in [Fe]-hydrogenase cofactor
Nat Commun, 6, 2015
|
|
3WV8
 
 | | ATP-bound HcgE from Methanothermobacter marburgensis | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Hmd co-occurring protein HcgE, SULFATE ION | | Authors: | Fujishiro, T, Ermler, U, Shima, S. | | Deposit date: | 2014-05-16 | | Release date: | 2015-04-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Protein-pyridinol thioester precursor for biosynthesis of the organometallic acyl-iron ligand in [Fe]-hydrogenase cofactor
Nat Commun, 6, 2015
|
|
3WB2
 
 | | HcgB from Methanocaldococcus jannaschii in complex with the guanylyl-pyridinol product in a model reaction of [Fe]-hydrogenase cofactor biosynthesis | | Descriptor: | 5'-O-[(R)-[(3,6-dimethyl-2-oxo-1,2-dihydropyridin-4-yl)oxy](hydroxy)phosphoryl]guanosine, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Fujishiro, T, Ermler, U, Shima, S. | | Deposit date: | 2013-05-11 | | Release date: | 2014-02-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Identification of the HcgB enzyme in [Fe]-hydrogenase-cofactor biosynthesis.
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
3WSE
 
 | | Reduced HcgD from Methanocaldococcus jannaschii | | Descriptor: | FE (II) ION, PHOSPHATE ION, Putative GTP cyclohydrolase 1 type 2 | | Authors: | Fujishiro, T, Ermler, U, Shima, S. | | Deposit date: | 2014-03-13 | | Release date: | 2014-08-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A possible iron delivery function of the dinuclear iron center of HcgD in [Fe]-hydrogenase cofactor biosynthesis
Febs Lett., 588, 2014
|
|
3V05
 
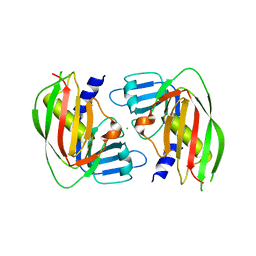 | | 2.4 Angstrom Crystal Structure of Superantigen-like Protein from Staphylococcus aureus. | | Descriptor: | CHLORIDE ION, Superantigen-like Protein | | Authors: | Minasov, G, Halavaty, A, Shuvalova, L, Filippova, E.V, Dubrovska, I, Winsor, J, Bagnoli, F, Falugi, F, Bottomley, M, Grandi, G, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-12-07 | | Release date: | 2011-12-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | 2.4 Angstrom Crystal Structure of Superantigen-like Protein from Staphylococcus aureus.
TO BE PUBLISHED
|
|
3WSF
 
 | |
3WVB
 
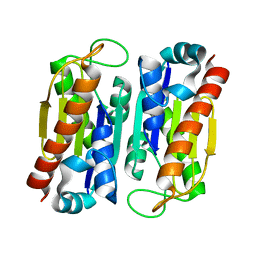 | | HcgF from Methanocaldococcus jannaschii | | Descriptor: | UPF0254 protein MJ1251 | | Authors: | Fujishiro, T, Ermler, U, Shima, S. | | Deposit date: | 2014-05-16 | | Release date: | 2015-04-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Protein-pyridinol thioester precursor for biosynthesis of the organometallic acyl-iron ligand in [Fe]-hydrogenase cofactor
Nat Commun, 6, 2015
|
|
3WVC
 
 | | Guanylylpyridinol (GP)-bound HcgF from Methanocaldococcus jannaschii | | Descriptor: | 5'-O-[(R)-hydroxy{[2-hydroxy-3,5-dimethyl-6-(2-oxo-2-sulfanylethyl)pyridin-4-yl]oxy}phosphoryl]guanosine, 5'-O-[(S)-{[2-(carboxymethyl)-6-hydroxy-3,5-dimethylpyridin-4-yl]oxy}(hydroxy)phosphoryl]guanosine, SULFATE ION, ... | | Authors: | Fujishiro, T, Ermler, U, Shima, S. | | Deposit date: | 2014-05-16 | | Release date: | 2015-04-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Protein-pyridinol thioester precursor for biosynthesis of the organometallic acyl-iron ligand in [Fe]-hydrogenase cofactor
Nat Commun, 6, 2015
|
|
3WVA
 
 | |
3WSH
 
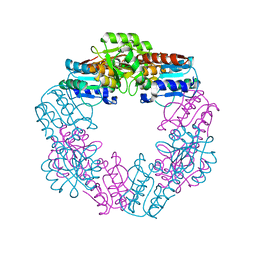 | | EDTA-treated, oxidized HcgD from Methanocaldococcus jannaschii | | Descriptor: | FE (III) ION, Putative GTP cyclohydrolase 1 type 2, SULFATE ION | | Authors: | Fujishiro, T, Ermler, U, Shima, S. | | Deposit date: | 2014-03-13 | | Release date: | 2014-08-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A possible iron delivery function of the dinuclear iron center of HcgD in [Fe]-hydrogenase cofactor biosynthesis
Febs Lett., 588, 2014
|
|
1D6R
 
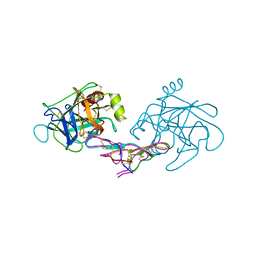 | | CRYSTAL STRUCTURE OF CANCER CHEMOPREVENTIVE BOWMAN-BIRK INHIBITOR IN TERNARY COMPLEX WITH BOVINE TRYPSIN AT 2.3 A RESOLUTION. STRUCTURAL BASIS OF JANUS-FACED SERINE PROTEASE INHIBITOR SPECIFICITY | | Descriptor: | BOWMAN-BIRK PROTEINASE INHIBITOR PRECURSOR, TRYPSINOGEN | | Authors: | Koepke, J, Ermler, U, Wenzl, G, Flecker, P. | | Deposit date: | 1999-10-15 | | Release date: | 2000-05-05 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of cancer chemopreventive Bowman-Birk inhibitor in ternary complex with bovine trypsin at 2.3 A resolution. Structural basis of Janus-faced serine protease inhibitor specificity.
J.Mol.Biol., 298, 2000
|
|
1D8B
 
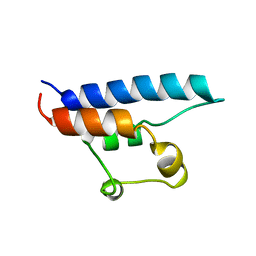 | | NMR STRUCTURE OF THE HRDC DOMAIN FROM SACCHAROMYCES CEREVISIAE RECQ HELICASE | | Descriptor: | SGS1 RECQ HELICASE | | Authors: | Liu, Z, Macias, M.J, Bottomley, M.J, Stier, G, Linge, J.P, Nilges, M, Bork, P, Sattler, M. | | Deposit date: | 1999-10-21 | | Release date: | 2000-01-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The three-dimensional structure of the HRDC domain and implications for the Werner and Bloom syndrome proteins.
Structure Fold.Des., 7, 1999
|
|
3WSI
 
 | | EDTA-treated, reduced HcgD from Methanocaldococcus jannaschii | | Descriptor: | FE (II) ION, PHOSPHATE ION, Putative GTP cyclohydrolase 1 type 2 | | Authors: | Fujishiro, T, Ermler, U, Shima, S. | | Deposit date: | 2014-03-13 | | Release date: | 2014-08-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A possible iron delivery function of the dinuclear iron center of HcgD in [Fe]-hydrogenase cofactor biosynthesis
Febs Lett., 588, 2014
|
|
3WSG
 
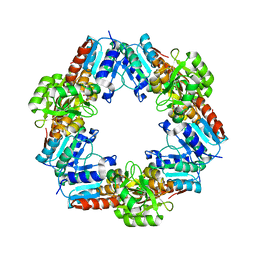 | |
3SQG
 
 | | Crystal structure of a methyl-coenzyme M reductase purified from Black Sea mats | | Descriptor: | (17[2]S)-17[2]-methylthio-coenzyme F43, 1-THIOETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Shima, S, Krueger, M, Weinert, T, Demmer, U, Thauer, R.K, Ermler, U. | | Deposit date: | 2011-07-05 | | Release date: | 2011-11-30 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of a methyl-coenzyme M reductase from Black Sea mats that oxidize methane anaerobically.
Nature, 481, 2011
|
|
3T41
 
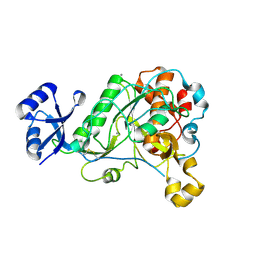 | | 1.95 Angstrom Resolution Crystal Structure of Epidermin Leader Peptide Processing Serine Protease (EpiP) S393A Mutant from Staphylococcus aureus | | Descriptor: | CALCIUM ION, CHLORIDE ION, Epidermin leader peptide processing serine protease EpiP | | Authors: | Minasov, G, Kuhn, M, Ruan, J, Halavaty, A, Shuvalova, L, Dubrovska, I, Winsor, J, Bagnoli, F, Falugi, F, Bottomley, M, Grandi, G, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-07-25 | | Release date: | 2011-08-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | 1.95 Angstrom Resolution Crystal Structure of Epidermin Leader Peptide Processing Serine Protease (EpiP) S393A Mutant from Staphylococcus aureus.
TO BE PUBLISHED
|
|
3SSV
 
 | |
3SVD
 
 | | Engineered medium-affinity halide-binding protein derived from YFP: bromide complex | | Descriptor: | 1,2-ETHANEDIOL, BROMIDE ION, Green fluorescent protein | | Authors: | Wang, W, Grimley, J.S, Beese, L.S, Hellinga, H.W. | | Deposit date: | 2011-07-12 | | Release date: | 2012-07-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Determination of engineered chloride-binding site structures in fluorescent proteins reveals principles of halide recognition
To be Published
|
|
4HW8
 
 | | 2.25 Angstrom Structure of the Extracellular Solute-binding Protein from Staphylococcus aureus in complex with Maltose. | | Descriptor: | Bacterial extracellular solute-binding protein, putative, CHLORIDE ION, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Winsor, J, Bagnoli, F, Falugi, F, Bottomley, M, Grandi, G, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-11-07 | | Release date: | 2012-11-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.251 Å) | | Cite: | 2.25 Angstrom Structure of the Extracellular Solute-binding Protein from Staphylococcus aureus in complex with Maltose.
TO BE PUBLISHED
|
|
4JJF
 
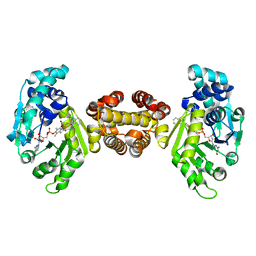 | | Crystal structure of FE-hydrogenase from methanothermobacter marburgensis in complex with 2-naphthylisocyanide | | Descriptor: | 5,10-methenyltetrahydromethanopterin hydrogenase, N-(naphthalen-2-yl)methanimine, iron-guanylyl pyridinol cofactor | | Authors: | Tamura, H, Warkentin, E, Ermler, U, Shima, S. | | Deposit date: | 2013-03-07 | | Release date: | 2013-08-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of [fe]-hydrogenase in complex with inhibitory isocyanides: implications for the h2 -activation site.
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
4JJG
 
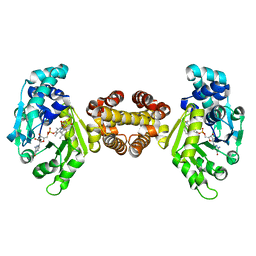 | | Crystal structure of FE-hydrogenase from methanothermobacter marburgensis in complex with toluenesulfonylmethylisocyanide | | Descriptor: | 5,10-methenyltetrahydromethanopterin hydrogenase, N-methyl-1-[(4-methylbenzyl)sulfonyl]methanamine, iron-guanylyl pyridinol cofactor | | Authors: | Tamura, H, Warkentin, E, Ermler, U, Shima, S. | | Deposit date: | 2013-03-07 | | Release date: | 2013-08-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of [fe]-hydrogenase in complex with inhibitory isocyanides: implications for the h2 -activation site.
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
4KPU
 
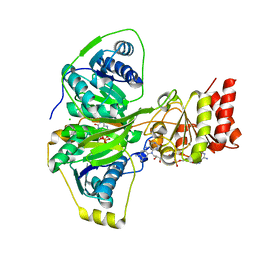 | | Electron transferring flavoprotein of Acidaminococcus fermentans: Towards a mechanism of flavin-based electron bifurcation | | Descriptor: | CHLORIDE ION, Electron transfer flavoprotein alpha subunit, Electron transfer flavoprotein alpha/beta-subunit, ... | | Authors: | Mowafy, A.M, Chowdhury, N.P, Demmer, J, Upadhyay, V, Kolzer, S, Jayamani, E, Kahnt, J, Demmer, U, Ermler, U, Buckel, W. | | Deposit date: | 2013-05-14 | | Release date: | 2014-01-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Studies on the Mechanism of Electron Bifurcation Catalyzed by Electron Transferring Flavoprotein (Etf) and Butyryl-CoA Dehydrogenase (Bcd) of Acidaminococcus fermentans.
J.Biol.Chem., 289, 2014
|
|
4L2I
 
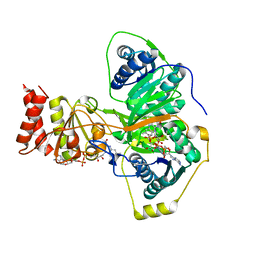 | | Electron transferring flavoprotein of Acidaminococcus fermentans: Towards a mechanism of flavin-based electron bifurcation | | Descriptor: | CHLORIDE ION, Electron transfer flavoprotein alpha subunit, Electron transfer flavoprotein alpha/beta-subunit, ... | | Authors: | Mowafy, A.M, Chowdhury, N.P, Demmer, J, Upadhyay, V, Kolzer, S, Jayamani, E, Kahnt, J, Demmer, U, Ermler, U, Buckel, W. | | Deposit date: | 2013-06-04 | | Release date: | 2014-01-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Studies on the Mechanism of Electron Bifurcation Catalyzed by Electron Transferring Flavoprotein (Etf) and Butyryl-CoA Dehydrogenase (Bcd) of Acidaminococcus fermentans.
J.Biol.Chem., 289, 2014
|
|
4L1F
 
 | | Electron transferring flavoprotein of Acidaminococcus fermentans: Towards a mechanism of flavin-based electron bifurcation | | Descriptor: | 1,3-PROPANDIOL, Acyl-CoA dehydrogenase domain protein, COENZYME A PERSULFIDE, ... | | Authors: | Mowafy, A.M, Chowdhury, N.P, Demmer, J, Upadhyay, V, Kolzer, S, Jayamani, E, Kahnt, J, Demmer, U, Ermler, U, Buckel, W. | | Deposit date: | 2013-06-03 | | Release date: | 2014-01-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Studies on the Mechanism of Electron Bifurcation Catalyzed by Electron Transferring Flavoprotein (Etf) and Butyryl-CoA Dehydrogenase (Bcd) of Acidaminococcus fermentans.
J.Biol.Chem., 289, 2014
|
|
