4B9L
 
 | | Structure of the high fidelity DNA polymerase I with the oxidative formamidopyrimidine-dA DNA lesion in the pre-insertion site. | | Descriptor: | 5'-D(*CP*AP*GP*FAX*AP*GP*AP*GP*TP*CP*AP*GP*GP*CP*TP)-3', 5'-D(*GP*CP*CP*TP*GP*AP*CP*TP*CP*TP)-3', DNA POLYMERASE I, ... | | Authors: | Gehrke, T.H, Lischke, U, Arnold, S, Schneider, S, Carell, T. | | Deposit date: | 2012-09-05 | | Release date: | 2013-04-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Unexpected Non-Hoogsteen-Based Mutagenicity Mechanism of Fapy-DNA Lesions.
Nat.Chem.Biol., 9, 2013
|
|
4AZ2
 
 | | Human thrombin - inhibitor complex | | Descriptor: | (R)-N-((S)-1-CARBAMIMIDOYL-PIPERIDIN-3-YLMETHYL)-2-(NAPHTHALENE-2-SULFONYLAMINO)-3-PHENYL-PROPIONAMIDE, 2-acetamido-2-deoxy-beta-D-glucopyranose, HIRUDIN-3A', ... | | Authors: | Banner, D.W, D'Arcy, A, Winkler, F.K, Hilpert, K. | | Deposit date: | 2012-06-22 | | Release date: | 2012-08-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Design and Synthesis of Potent and Highly Selective Thrombin Inhibitors.
J.Med.Chem., 37, 1994
|
|
293D
 
 | | INTERACTION BETWEEN THE LEFT-HANDED Z-DNA AND POLYAMINE-2: THE CRYSTAL STRUCTURE OF THE D(CG)3 AND SPERMIDINE COMPLEX | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*CP*G)-3'), MAGNESIUM ION, SODIUM ION, ... | | Authors: | Ohishi, H, Nakanishi, I, Inubushi, K, Van Der Marel, G.A, Van Boom, J.H, Rich, A, Wang, A.H.-J, Hakoshima, T, Tomita, K. | | Deposit date: | 1996-10-09 | | Release date: | 1996-12-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Interaction between the left-handed Z-DNA and polyamine-2. The crystal structure of the d(CG)3 and spermidine complex.
FEBS Lett., 391, 1996
|
|
4B2T
 
 | | The crystal structures of the eukaryotic chaperonin CCT reveal its functional partitioning | | Descriptor: | T-COMPLEX PROTEIN 1 SUBUNIT ALPHA, T-COMPLEX PROTEIN 1 SUBUNIT BETA, T-COMPLEX PROTEIN 1 SUBUNIT DELTA, ... | | Authors: | Kalisman, N, Schroeder, G.F, Levitt, M. | | Deposit date: | 2012-07-17 | | Release date: | 2013-03-20 | | Last modified: | 2019-11-06 | | Method: | X-RAY DIFFRACTION (5.5 Å) | | Cite: | The Crystal Structures of the Eukaryotic Chaperonin Cct Reveal its Functional Partitioning
Structure, 21, 2013
|
|
4AWJ
 
 | | pVHL:EloB:EloC complex, in complex with capped Hydroxyproline | | Descriptor: | (4R)-1-acetyl-4-hydroxy-N-methyl-L-prolinamide, ACETATE ION, ACETIC ACID, ... | | Authors: | Van Molle, I, Thomann, A, Buckley, D.L, So, E.C, Lang, S, Crews, C.M, Ciulli, A. | | Deposit date: | 2012-06-04 | | Release date: | 2012-11-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Dissecting Fragment-Based Lead Discovery at the Von Hippel-Lindau Protein:Hypoxia Inducible Factor 1Alpha Protein-Protein Interface.
Chem.Biol., 19, 2012
|
|
4AXV
 
 | | Biochemical and structural characterization of the MpaA amidase as part of a conserved scavenging pathway for peptidoglycan derived peptides in gamma-proteobacteria | | Descriptor: | 1,2-ETHANEDIOL, MPAA, ZINC ION | | Authors: | Maqbool, A, Herve, M, Mengin-Lecreulx, D, Dodson, E, Wilkinson, A.J, Thomas, G.H. | | Deposit date: | 2012-06-14 | | Release date: | 2012-09-26 | | Last modified: | 2020-03-04 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Mpaa is a Murein-Tripeptide-Specific Zinc Carboxypeptidase that Functions as Part of a Catabolic Pathway for Peptidoglycan Derived Peptides in Gamma-Proteobacteria.
Biochem.J., 448, 2012
|
|
4B73
 
 | | Discovery of an allosteric mechanism for the regulation of HCV NS3 protein function | | Descriptor: | (2S)-4-amino-N-[(1R)-1-(4-chloro-2-fluoro-3-phenoxyphenyl)propyl]-4-oxobutan-2-aminium, NON-STRUCTURAL PROTEIN 4A, SERINE PROTEASE NS3 | | Authors: | Saalau-Bethell, S.M, Woodhead, A.J, Chessari, G, Carr, M.G, Coyle, J, Graham, B, Hiscock, S.D, Murray, C.W, Pathuri, P, Rich, S.J, Richardson, C.J, Williams, P.A, Jhoti, H. | | Deposit date: | 2012-08-16 | | Release date: | 2012-10-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of an Allosteric Mechanism for the Regulation of Hcv Ns3 Protein Function.
Nat.Chem.Biol., 8, 2012
|
|
2DVN
 
 | |
5V34
 
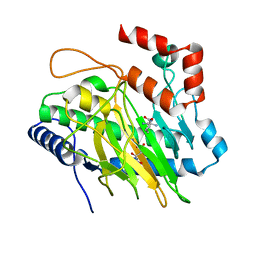 | | Ethylene forming enzyme in complex with manganese, malic acid and L-arginine | | Descriptor: | 2-oxoglutarate-dependent ethylene/succinate-forming enzyme, ARGININE, D-MALATE, ... | | Authors: | Fellner, M, Martinez, S, Hu, J, Hausinger, R.P. | | Deposit date: | 2017-03-06 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Structures and Mechanisms of the Non-Heme Fe(II)- and 2-Oxoglutarate-Dependent Ethylene-Forming Enzyme: Substrate Binding Creates a Twist.
J. Am. Chem. Soc., 139, 2017
|
|
2DW1
 
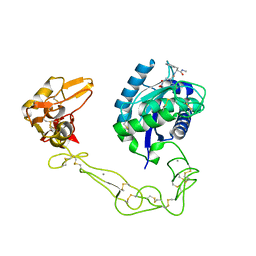 | | Crystal structure of VAP2 from Crotalus atrox venom (Form 2-2 crystal) | | Descriptor: | 3-(N-HYDROXYCARBOXAMIDO)-2-ISOBUTYLPROPANOYL-TRP-METHYLAMIDE, CALCIUM ION, Catrocollastatin, ... | | Authors: | Takeda, S, Igarashi, T, Araki, S. | | Deposit date: | 2006-08-02 | | Release date: | 2007-07-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of catrocollastatin/VAP2B reveal a dynamic, modular architecture of ADAM/adamalysin/reprolysin family proteins
Febs Lett., 581, 2007
|
|
5V3J
 
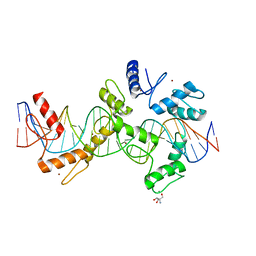 | | mouseZFP568-ZnF1-10 in complex with DNA | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DNA (26-MER), MAGNESIUM ION, ... | | Authors: | Patel, A, Cheng, X. | | Deposit date: | 2017-03-07 | | Release date: | 2018-03-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.064 Å) | | Cite: | DNA Conformation Induces Adaptable Binding by Tandem Zinc Finger Proteins.
Cell, 173, 2018
|
|
2DCQ
 
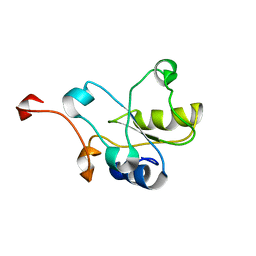 | |
5V5V
 
 | | Complex of NLGN2 with MDGA1 Ig1-Ig2 | | Descriptor: | MAM domain-containing glycosylphosphatidylinositol anchor protein 1, Neuroligin-2 | | Authors: | Gangwar, S.P, Machius, M, Rudenko, G. | | Deposit date: | 2017-03-15 | | Release date: | 2017-07-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (4.11 Å) | | Cite: | Molecular Mechanism of MDGA1: Regulation of Neuroligin 2:Neurexin Trans-synaptic Bridges.
Neuron, 94, 2017
|
|
4B8V
 
 | | Cladosporium fulvum LysM effector Ecp6 in complex with a beta-1,4- linked N-acetyl-D-glucosamine tetramer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, ... | | Authors: | Saleem-Batcha, R, Sanchez-Vallet, A, Hansen, G, Thomma, B.P.H.J, Mesters, J.R. | | Deposit date: | 2012-08-30 | | Release date: | 2013-07-17 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Fungal Effector Ecp6 Outcompetes Host Immune Receptor for Chitin Binding Through Intrachain Lysm Dimerization
Elife, 2, 2013
|
|
1ZV4
 
 | | Structure of the Regulator of G-Protein Signaling 17 (RGSZ2) | | Descriptor: | Regulator of G-protein signaling 17 | | Authors: | Schoch, G.A, Jansson, A, Elkins, J.M, Haroniti, A, Niesen, F.H, Bunkoczi, G, Lee, W.H, Turnbull, A.P, Yang, X, Sundstrom, M, Arrowsmith, C, Edwards, A, Marsden, B, Gileadi, O, Ball, L, von Delft, F, Doyle, D.A, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-06-01 | | Release date: | 2005-06-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural diversity in the RGS domain and its interaction with heterotrimeric G protein alpha-subunits.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
4ATU
 
 | | Human doublecortin N-DC repeat plus linker, and tubulin (2XRP) docked into an 8A cryo-EM map of doublecortin-stabilised microtubules reconstructed in absence of kinesin | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, NEURONAL MIGRATION PROTEIN DOUBLECORTIN, ... | | Authors: | Liu, J.S, Schubert, C.R, Fu, X, Fourniol, F.J, Jaiswal, J.K, Houdusse, A, Stultz, C.M, Moores, C.A, Walsh, C.A. | | Deposit date: | 2012-05-09 | | Release date: | 2012-09-26 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (8.3 Å) | | Cite: | Molecular Basis for Specific Regulation of Neuronal Kinesin- 3 Motors by Doublecortin Family Proteins.
Mol.Cell, 47, 2012
|
|
1ZVL
 
 | | Rat Neuronal Nitric Oxide Synthase Oxygenase Domain complexed with natural substrate L-Arg. | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ARGININE, Nitric-oxide synthase, ... | | Authors: | Matter, H, Kumar, H.S, Fedorov, R, Frey, A, Kotsonis, P, Hartmann, E, Frohlich, L.G, Reif, A, Pfleiderer, W, Scheurer, P, Ghosh, D.K, Schlichting, I, Schmidt, H.H. | | Deposit date: | 2005-06-02 | | Release date: | 2005-08-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Analysis of Isoform-Specific Inhibitors Targeting the Tetrahydrobiopterin Binding Site of Human Nitric Oxide Synthases.
J.Med.Chem., 48, 2005
|
|
2DFS
 
 | | 3-D structure of Myosin-V inhibited state | | Descriptor: | Calmodulin, Myosin-5A | | Authors: | Liu, J, Taylor, D.W, Krementsova, E.B, Trybus, K.M, Taylor, K.A. | | Deposit date: | 2006-03-03 | | Release date: | 2006-04-25 | | Last modified: | 2024-03-13 | | Method: | ELECTRON CRYSTALLOGRAPHY (24 Å) | | Cite: | Three-dimensional structure of the myosin V inhibited state by cryoelectron tomography
Nature, 442, 2006
|
|
4AW8
 
 | | X-ray structure of ZinT from Salmonella enterica in complex with zinc ion and PEG | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, Metal-binding protein ZinT, SODIUM ION, ... | | Authors: | Alaleona, F, Ilari, A, Battistoni, A, Petrarca, P, Chiancone, E. | | Deposit date: | 2012-06-01 | | Release date: | 2013-06-19 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Salmonella Enterica Zint Structure, Zinc Affinity and Interaction with the High-Affinity Uptake Protein Znua Provide Insight Into the Management of Periplasmic Zinc.
Biochim.Biophys.Acta, 1840, 2014
|
|
4AVU
 
 | | Crystal structure of human tankyrase 2 in complex with 6(5H)- phenanthridinone | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, SULFATE ION, ... | | Authors: | Haikarainen, T, Narwal, M, Lehtio, L. | | Deposit date: | 2012-05-29 | | Release date: | 2012-06-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Evaluation and Structural Basis for the Inhibition of Tankyrases by Parp Inhibitors
Acs Med.Chem.Lett., 5, 2014
|
|
5V2U
 
 | | Ethylene forming enzyme apo form | | Descriptor: | 2-oxoglutarate-dependent ethylene/succinate-forming enzyme | | Authors: | Fellner, M, Martinez, S, Hu, J, Hausinger, R.P. | | Deposit date: | 2017-03-06 | | Release date: | 2017-08-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.058 Å) | | Cite: | Structures and Mechanisms of the Non-Heme Fe(II)- and 2-Oxoglutarate-Dependent Ethylene-Forming Enzyme: Substrate Binding Creates a Twist.
J. Am. Chem. Soc., 139, 2017
|
|
5V32
 
 | | Ethylene forming enzyme in complex with manganese and malic acid | | Descriptor: | 2-oxoglutarate-dependent ethylene/succinate-forming enzyme, D-MALATE, MANGANESE (II) ION | | Authors: | Fellner, M, Martinez, S, Hu, J, Hausinger, R.P. | | Deposit date: | 2017-03-06 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.486 Å) | | Cite: | Structures and Mechanisms of the Non-Heme Fe(II)- and 2-Oxoglutarate-Dependent Ethylene-Forming Enzyme: Substrate Binding Creates a Twist.
J. Am. Chem. Soc., 139, 2017
|
|
2DJ2
 
 | | The solution structure of the second thioredoxin domain of mouse Protein disulfide-isomerase A4 | | Descriptor: | Protein disulfide-isomerase A4 | | Authors: | Tochio, N, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-03-30 | | Release date: | 2006-09-30 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the second thioredoxin domain of mouse Protein disulfide-isomerase A4
To be Published
|
|
5V4Q
 
 | | Crystal Structure of human GGT1 in complex with DON | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5,5-dihydroxy-L-norleucine, CHLORIDE ION, ... | | Authors: | Terzyan, S, Hanigan, M. | | Deposit date: | 2017-03-10 | | Release date: | 2017-04-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of 6-diazo-5-oxo-norleucine-bound human gamma-glutamyl transpeptidase 1, a novel mechanism of inactivation.
Protein Sci., 26, 2017
|
|
1ZZS
 
 | | Bovine eNOS N368D single mutant with L-N(omega)-Nitroarginine-(4R)-Amino-L-Proline Amide Bound | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, GLYCEROL, ... | | Authors: | Li, H, Flinspach, M.L, Igarashi, J, Jamal, J, Yang, W, Gomez-Vidal, J.A, Litzinger, E.A, Silverman, R.B, Poulos, T.L. | | Deposit date: | 2005-06-14 | | Release date: | 2005-12-06 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Exploring the Binding Conformations of Bulkier Dipeptide Amide Inhibitors in Constitutive Nitric Oxide Synthases.
Biochemistry, 44, 2005
|
|
