2CIC
 
 | | THE CRYSTAL STRUCTURE OF A COMPLEX OF CAMPYLOBACTER JEJUNI DUTPASE WITH SUBSTRATE ANALOGUE DUPNHPP | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, DEOXYURIDINE 5'-TRIPHOSPHATE NUCLEOTIDE HYDROLASE, MAGNESIUM ION | | Authors: | Moroz, O.V, Harkiolaki, M, Gonzalez-Pacanowska, D, Wilson, K.S. | | Deposit date: | 2006-03-17 | | Release date: | 2007-03-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Crystal Structure of the Leishmania Major Deoxyuridine Triphosphate Nucleotidohydrolase in Complex with Nucleotide Analogues, Dump, and Deoxyuridine.
J.Biol.Chem., 286, 2011
|
|
6OW7
 
 | | X-ray Structure of Polypeptide Deformylase with a Piperazic Acid | | Descriptor: | (3S)-2-{(2R)-2-(cyclopentylmethyl)-3-[formyl(hydroxy)amino]propanoyl}-N-(pyridin-2-yl)hexahydropyridazine-3-carboxamide, NICKEL (II) ION, Peptide deformylase, ... | | Authors: | Campobasso, N, Spletstoser, J, Ward, P. | | Deposit date: | 2019-05-09 | | Release date: | 2019-06-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Discovery of piperazic acid peptide deformylase inhibitors with in vivo activity for respiratory tract and skin infections.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
1H97
 
 | | Trematode hemoglobin from Paramphistomum epiclitum | | Descriptor: | Globin-3, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Pesce, A, Dewilde, S, Kiger, L, Milani, M, Ascenzi, P, Marden, M.C, Van Hauwaert, M.L, Vanfleteren, J, Moens, L, Bolognesi, M. | | Deposit date: | 2001-03-02 | | Release date: | 2001-06-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Very High Resolution Structure of a Trematode Hemoglobin Displaying a Tyrb10-Tyre7 Heme Distal Residue Pair and High Oxygen Affinity
J.Mol.Biol., 309, 2001
|
|
8DTJ
 
 | | Human NAMPT in complex with small molecule activator ZN-43-S | | Descriptor: | (3S)-N-[(1-benzothiophen-5-yl)methyl]-1-{2-[4-(trifluoromethyl)phenyl]-2H-pyrazolo[3,4-d]pyrimidin-4-yl}piperidine-3-carboxamide, CHLORIDE ION, GLYCEROL, ... | | Authors: | Ratia, K, Xiong, R, Shen, Z, Thatcher, G.R. | | Deposit date: | 2022-07-25 | | Release date: | 2023-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.118 Å) | | Cite: | Mechanism of Allosteric Modulation of Nicotinamide Phosphoribosyltransferase to Elevate Cellular NAD.
Biochemistry, 62, 2023
|
|
8DSD
 
 | | Human NAMPT in complex with substrate NAM and small molecule activator NP-A1-S | | Descriptor: | (3S)-1-[2-(4-methylphenyl)-2H-pyrazolo[3,4-d]pyrimidin-4-yl]-N-{[4-(methylsulfanyl)phenyl]methyl}piperidine-3-carboxamide, CHLORIDE ION, GLYCEROL, ... | | Authors: | Ratia, K, Xiong, R, Shen, Z, Thatcher, G.R. | | Deposit date: | 2022-07-22 | | Release date: | 2023-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.429 Å) | | Cite: | Mechanism of Allosteric Modulation of Nicotinamide Phosphoribosyltransferase to Elevate Cellular NAD.
Biochemistry, 62, 2023
|
|
8DSH
 
 | | Human NAMPT in complex with quercitrin and AMPcP | | Descriptor: | 2-(3,4-dihydroxyphenyl)-5,7-dihydroxy-4-oxo-4H-chromen-3-yl 6-deoxy-alpha-L-mannopyranoside, Nicotinamide phosphoribosyltransferase, PHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER | | Authors: | Ratia, K, Xiong, R, Shen, Z, Thatcher, G.R. | | Deposit date: | 2022-07-22 | | Release date: | 2023-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mechanism of Allosteric Modulation of Nicotinamide Phosphoribosyltransferase to Elevate Cellular NAD.
Biochemistry, 62, 2023
|
|
8DSI
 
 | | Human NAMPT in complex with substrate NAM | | Descriptor: | CHLORIDE ION, GLYCEROL, NICOTINAMIDE, ... | | Authors: | Ratia, K, Xiong, R, Shen, Z, Thatcher, G.R. | | Deposit date: | 2022-07-22 | | Release date: | 2023-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Mechanism of Allosteric Modulation of Nicotinamide Phosphoribosyltransferase to Elevate Cellular NAD.
Biochemistry, 62, 2023
|
|
8DSC
 
 | | Human NAMPT in complex with substrate NAM and small molecule activator NP-A1-R | | Descriptor: | (3R)-1-[2-(4-methylphenyl)-2H-pyrazolo[3,4-d]pyrimidin-4-yl]-N-{[4-(methylsulfanyl)phenyl]methyl}piperidine-3-carboxamide, CHLORIDE ION, GLYCEROL, ... | | Authors: | Ratia, K, Xiong, R, Shen, Z, Thatcher, G.R. | | Deposit date: | 2022-07-22 | | Release date: | 2023-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.321 Å) | | Cite: | Mechanism of Allosteric Modulation of Nicotinamide Phosphoribosyltransferase to Elevate Cellular NAD.
Biochemistry, 62, 2023
|
|
8DSE
 
 | | Human NAMPT in complex with substrate NAM and activator quercitrin | | Descriptor: | 2-(3,4-dihydroxyphenyl)-5,7-dihydroxy-4-oxo-4H-chromen-3-yl 6-deoxy-alpha-L-mannopyranoside, CHLORIDE ION, NICOTINAMIDE, ... | | Authors: | Ratia, K, Xiong, R, Shen, Z, Thatcher, G.R. | | Deposit date: | 2022-07-22 | | Release date: | 2023-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.428 Å) | | Cite: | Mechanism of Allosteric Modulation of Nicotinamide Phosphoribosyltransferase to Elevate Cellular NAD.
Biochemistry, 62, 2023
|
|
5AZ4
 
 | |
3Q3V
 
 | | Crystal structure of Phosphoglycerate Kinase from Campylobacter jejuni. | | Descriptor: | FORMIC ACID, POTASSIUM ION, Phosphoglycerate kinase, ... | | Authors: | Filippova, E.V, Wawrzak, Z, Onopriyenko, O, Edwards, A, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-12-22 | | Release date: | 2011-01-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.145 Å) | | Cite: | Crystal structures of putative phosphoglycerate kinases from B. anthracis and C. jejuni.
J.Struct.Funct.Genom., 13, 2012
|
|
6OW2
 
 | | X-ray Structure of Polypeptide Deformylase | | Descriptor: | (2R)-2-(cyclopentylmethyl)-N'-{5-fluoro-6-[(9aS)-hexahydropyrazino[2,1-c][1,4]oxazin-8(1H)-yl]-2-methylpyrimidin-4-yl}-3-[hydroxy(hydroxymethyl)amino]propanehydrazide, NICKEL (II) ION, Peptide deformylase | | Authors: | Campobasso, N, Spletstoser, J, Ward, P. | | Deposit date: | 2019-05-09 | | Release date: | 2019-06-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of piperazic acid peptide deformylase inhibitors with in vivo activity for respiratory tract and skin infections.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
3NZP
 
 | | Crystal Structure of the Biosynthetic Arginine decarboxylase SpeA from Campylobacter jejuni, Northeast Structural Genomics Consortium Target BR53 | | Descriptor: | Arginine decarboxylase, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | Authors: | Forouhar, F, Lew, S, Seetharaman, J, Sahdev, S, Xiao, R, Ciccosanti, C, Belote, R.L, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-07-16 | | Release date: | 2010-09-01 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of bacterial biosynthetic arginine decarboxylases.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
3KMG
 
 | |
8UKP
 
 | | cAMP-dependent protein kinase A catalytic domain in complex with voltage gated calcium channel peptide ternary complex 1 | | Descriptor: | ARG-GLY-PHE-LEU-ARG-SER-ALA-SER-LEU-GLY-ARG-ARG-ALA-SER-PHE-HIS-LEU, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Haji-Ghassemi, O, Van Petegem, F. | | Deposit date: | 2023-10-14 | | Release date: | 2024-12-11 | | Last modified: | 2025-01-01 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystallographic, kinetic, and calorimetric investigation of PKA interactions with L-type calcium channels and Rad GTPase.
J.Biol.Chem., 301, 2024
|
|
8UKO
 
 | | cAMP-dependent protein kinase A catalytic domain in complex with voltage gated calcium channel peptide ternary complex 2 | | Descriptor: | ARG-GLY-PHE-LEU-ARG-SER-ALA-SER-LEU-GLY-ARG-ARG-ALA-SER-PHE-HIS-LEU, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Haji-Ghassemi, O, Van Petegem, F. | | Deposit date: | 2023-10-14 | | Release date: | 2024-12-11 | | Last modified: | 2025-01-01 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Crystallographic, kinetic, and calorimetric investigation of PKA interactions with L-type calcium channels and Rad GTPase.
J.Biol.Chem., 301, 2024
|
|
5LWQ
 
 | | CeuE (H227L variant) a periplasmic protein from Campylobacter jejuni | | Descriptor: | BROMIDE ION, Enterochelin uptake periplasmic binding protein, SODIUM ION | | Authors: | Wilde, E.J, Blagova, E, Hughes, A, Raines, D.J, Moroz, O.V, Turkenburg, J.P, Duhme-Klair, A.-K, Wilson, K.S. | | Deposit date: | 2016-09-19 | | Release date: | 2017-04-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Interactions of the periplasmic binding protein CeuE with Fe(III) n-LICAM(4-) siderophore analogues of varied linker length.
Sci Rep, 7, 2017
|
|
5LWH
 
 | | CeuE (Y288F variant) a periplasmic protein from Campylobacter jejuni. | | Descriptor: | Enterochelin ABC transporter substrate-binding protein, ZINC ION | | Authors: | Wilde, E.J, Blagova, E, Hughes, A, Raines, D.J, Moroz, O.V, Turkenburg, J, Duhme-Klair, A.-K, Wilson, K.S. | | Deposit date: | 2016-09-16 | | Release date: | 2017-04-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Interactions of the periplasmic binding protein CeuE with Fe(III) n-LICAM(4-) siderophore analogues of varied linker length.
Sci Rep, 7, 2017
|
|
5M0W
 
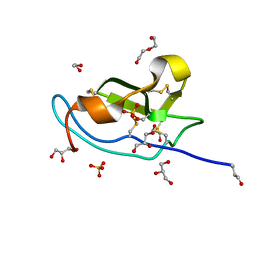 | | N-terminal domain of mouse Shisa 3 | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Lohkamp, B, Ojala, J.R.M. | | Deposit date: | 2016-10-06 | | Release date: | 2017-04-05 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Ab initio solution of macromolecular crystal structures without direct methods.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
1XNC
 
 | |
1Y9U
 
 | |
3NYU
 
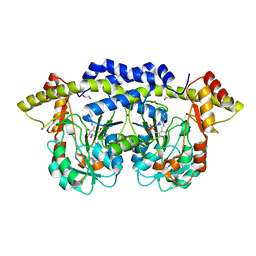 | |
4PMH
 
 | | The structure of rice weevil pectin methyl esterase | | Descriptor: | Pectinesterase | | Authors: | Stenkamp, R.E, Teller, D.C, Behnke, C.A, Reeck, G.R. | | Deposit date: | 2014-05-21 | | Release date: | 2014-11-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | The structure of rice weevil pectin methylesterase.
Acta Crystallogr.,Sect.F, 70, 2014
|
|
3STR
 
 | | Strep Peptide Deformylase with a time dependent thiazolidine hydroxamic acid | | Descriptor: | (4R)-3-(4-[4-(2-chlorophenyl)piperazin-1-yl]-6-{[2-methyl-6-(methylcarbamoyl)phenyl]amino}-1,3,5-triazin-2-yl)-N-[2-(hydroxyamino)-2-oxoethyl]-1,3-thiazolidine-4-carboxamide, GLYCEROL, NICKEL (II) ION, ... | | Authors: | Campobasso, N, Ward, P. | | Deposit date: | 2011-07-11 | | Release date: | 2011-07-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Understanding the origins of time-dependent inhibition by polypeptide deformylase inhibitors.
Biochemistry, 50, 2011
|
|
4R4I
 
 | | Structure of RPA70N in complex with 5-(4-((6-(5-carboxyfuran-2-yl)-1-thioxo-3,4-dihydroisoquinolin-2(1H)-yl)methyl)phenyl)-1-(3,4-dichlorophenyl)-1H-pyrazole-3-carboxylic acid | | Descriptor: | 5-(4-{[6-(5-carboxyfuran-2-yl)-1-thioxo-3,4-dihydroisoquinolin-2(1H)-yl]methyl}phenyl)-1-(3,4-dichlorophenyl)-1H-pyrazole-3-carboxylic acid, Replication protein A 70 kDa DNA-binding subunit | | Authors: | Feldkamp, M.D, Waterson, A.G, Kennedy, J.P, Patrone, J.D, Pelz, N.F, Frank, A.O, Vangamudi, B, Sousa-Fagundes, E.M, Rossanese, O.W, Fesik, S.W, Chazin, W.J. | | Deposit date: | 2014-08-19 | | Release date: | 2014-11-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Diphenylpyrazoles as replication protein a inhibitors.
ACS Med Chem Lett, 6, 2015
|
|
