8IB4
 
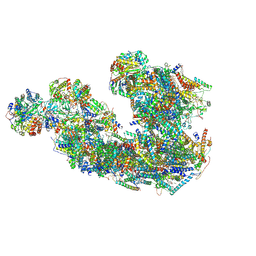 | | Respiratory complex CI:CIII2, type IA, Wild type mouse under cold temperature | | Descriptor: | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, ... | | Authors: | Shin, Y.-C, Liao, M. | | Deposit date: | 2023-02-09 | | Release date: | 2024-09-18 | | Last modified: | 2024-12-04 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural basis of respiratory complex adaptation to cold temperatures.
Cell, 187, 2024
|
|
8IB9
 
 | | Respiratory complex CI:CIII2, type IB, Wild type mouse under cold temperature | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 5-(3,7,11,15,19,23-HEXAMETHYL-TETRACOSA-2,6,10,14,18,22-HEXAENYL)-2,3-DIMETHOXY-6-METHYL-BENZENE-1,4-DIOL, ... | | Authors: | Shin, Y.-C, Liao, M. | | Deposit date: | 2023-02-10 | | Release date: | 2024-09-18 | | Last modified: | 2024-12-04 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural basis of respiratory complex adaptation to cold temperatures.
Cell, 187, 2024
|
|
5D8L
 
 | |
5QCD
 
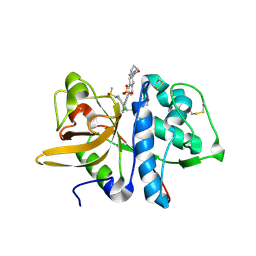 | | Crystal structure of human Cathepsin-S with bound ligand | | Descriptor: | Cathepsin S, SULFATE ION, {(3S)-7-[(2-chloro-5-{5-(methylsulfonyl)-1-[3-(morpholin-4-yl)propyl]-4,5,6,7-tetrahydro-1H-pyrazolo[4,3-c]pyridin-3-yl}phenyl)ethynyl]-1,2,3,4-tetrahydroisoquinolin-3-yl}(piperidin-1-yl)methanone | | Authors: | Bembenek, S.D, Ameriks, M.K, Mirzadegan, T, Yang, H, Shao, C, Burley, S.K. | | Deposit date: | 2017-08-04 | | Release date: | 2017-12-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of human Cathepsin-S with bound ligand
To be published
|
|
5QCI
 
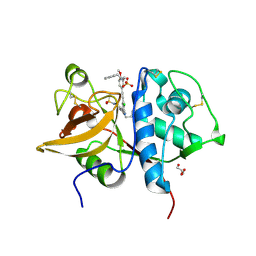 | | Crystal structure of human Cathepsin-S with bound ligand | | Descriptor: | Cathepsin S, GLYCEROL, N-benzyl-1-{5-[(2-chloro-5-{5-(methylsulfonyl)-1-[3-(morpholin-4-yl)propyl]-4,5,6,7-tetrahydro-1H-pyrazolo[4,3-c]pyridin-3-yl}phenyl)ethynyl]-2-methoxyphenyl}methanamine, ... | | Authors: | Bembenek, S.D, Ameriks, M.K, Mirzadegan, T, Yang, H, Shao, C, Burley, S.K. | | Deposit date: | 2017-08-04 | | Release date: | 2017-12-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.179 Å) | | Cite: | Crystal structure of human Cathepsin-S with bound ligand
To be published
|
|
5QCE
 
 | | Crystal structure of human Cathepsin-S with bound ligand | | Descriptor: | Cathepsin S, N-benzyl-1-{2-chloro-5-[(2-chloro-5-{5-(methylsulfonyl)-1-[3-(morpholin-4-yl)propyl]-4,5,6,7-tetrahydro-1H-pyrazolo[4,3-c]pyridin-3-yl}phenyl)ethynyl]phenyl}methanamine | | Authors: | Bembenek, S.D, Ameriks, M.K, Mirzadegan, T, Yang, H, Shao, C, Burley, S.K. | | Deposit date: | 2017-08-04 | | Release date: | 2017-12-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Crystal structure of human Cathepsin-S with bound ligand
To be published
|
|
5QBU
 
 | | Crystal structure of human Cathepsin-S with bound ligand | | Descriptor: | 1-[1-(3-{5-(1H-imidazole-5-carbonyl)-3-[4-(trifluoromethyl)phenyl]-4,5,6,7-tetrahydro-1H-pyrazolo[4,3-c]pyridin-1-yl}propyl)piperidin-4-yl]-3-methyl-1,3-dihydro-2H-benzimidazol-2-one, Cathepsin S | | Authors: | Bembenek, S.D, Ameriks, M.K, Mirzadegan, T, Yang, H, Shao, C, Burley, S.K. | | Deposit date: | 2017-08-04 | | Release date: | 2017-12-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Crystal structure of human Cathepsin-S with bound ligand
To be published
|
|
5QC5
 
 | | Crystal structure of human Cathepsin-S with bound ligand | | Descriptor: | 1-[5-{1-[3-(4-tert-butylpiperidin-1-yl)propyl]-5-(methylsulfonyl)-4,5,6,7-tetrahydro-1H-pyrazolo[4,3-c]pyridin-3-yl}-2-(trifluoromethyl)phenyl]-N-[(4-fluorophenyl)methyl]methanamine, Cathepsin S | | Authors: | Bembenek, S.D, Ameriks, M.K, Mirzadegan, T, Yang, H, Shao, C, Burley, S.K. | | Deposit date: | 2017-08-04 | | Release date: | 2017-12-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of human Cathepsin-S with bound ligand
To be published
|
|
9MO5
 
 | |
9MO8
 
 | |
9MOB
 
 | |
9QOB
 
 | | CRYSTAL STRUCTURE OF THE FIRST BROMODOMAIN OF HUMAN BRD4 IN COMPLEX with compound 4 | | Descriptor: | (5-phenyl-[1,3]thiazolo[2,3-c][1,2,4]triazol-3-yl)methanol, 1,2-ETHANEDIOL, Bromodomain-containing protein 4 | | Authors: | Krojer, T, Martinez-Cartro, M, Picaud, S, Filippakopoulos, P, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Barril, X, von Delft, F. | | Deposit date: | 2025-03-25 | | Release date: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | CRYSTAL STRUCTURE OF THE FIRST BROMODOMAIN OF HUMAN BRD4 IN COMPLEX with compound 2
To Be Published
|
|
9MOP
 
 | |
9MOL
 
 | |
9MOO
 
 | |
9MOM
 
 | |
9MOA
 
 | |
9MO6
 
 | |
9MO7
 
 | |
9MO9
 
 | |
9MZA
 
 | | Chemically Hijacked BCL6-TCIP3-p300 Complex | | Descriptor: | 1-{1-[5-({1-[5-chloro-4-({8-methoxy-1-methyl-3-[2-(methylamino)-2-oxoethoxy]-2-oxo-1,2-dihydroquinolin-6-yl}amino)pyrimidin-2-yl]piperidine-4-carbonyl}amino)pentanoyl]piperidin-4-yl}-3-[(6M)-7-(difluoromethyl)-6-(1-methyl-1H-pyrazol-4-yl)-3,4-dihydroquinolin-1(2H)-yl]-N-methyl-1,4,6,7-tetrahydro-5H-pyrazolo[4,3-c]pyridine-5-carboxamide, B-cell lymphoma 6 protein, Histone acetyltransferase p300 | | Authors: | Hinshaw, S.M, Gray, N.S, Nix, M.N, Gourisankar, S, Martinez, M, Crabtree, G.R. | | Deposit date: | 2025-01-22 | | Release date: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Bivalent Molecular Glue Linking Lysine Acetyltransferases to Oncogene-induced Cell Death.
Biorxiv, 2025
|
|
9MOI
 
 | |
9MOX
 
 | |
9MO4
 
 | |
9MOD
 
 | |
