6EDZ
 
 | |
6IV9
 
 | | the Cas13d binary complex | | Descriptor: | Cas13d, MAGNESIUM ION, crRNA (50-MER) | | Authors: | Zhang, B, Ye, Y.M, Ye, W.W, OuYang, S.Y. | | Deposit date: | 2018-12-02 | | Release date: | 2019-06-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Two HEPN domains dictate CRISPR RNA maturation and target cleavage in Cas13d.
Nat Commun, 10, 2019
|
|
6EEY
 
 | |
6I4V
 
 | |
6E4C
 
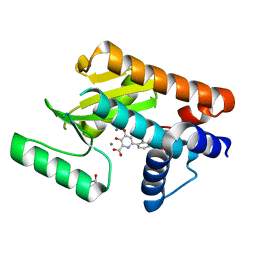 | | The N-terminal domain of PA endonuclease from the influenza H1N1 virus in complex with 3-hydroxy-6-(2-methyl-4-(5-oxo-4,5-dihydro-1,2,4-oxadiazol-3-yl)phenyl)-4-oxo-1,4-dihydropyridine-2-carboxylic acid | | Descriptor: | 1,2-ETHANEDIOL, 3-hydroxy-6-[2-methyl-4-(5-oxo-4,5-dihydro-1,2,4-oxadiazol-3-yl)phenyl]-4-oxo-1,4-dihydropyridine-2-carboxylic acid, MANGANESE (II) ION, ... | | Authors: | Morrison, C.N, Dick, B.L, Credille, C.V, Cohen, S.M. | | Deposit date: | 2018-07-17 | | Release date: | 2019-07-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | SAR Exploration of Tight-Binding Inhibitors of Influenza Virus PA Endonuclease.
J.Med.Chem., 62, 2019
|
|
6EF5
 
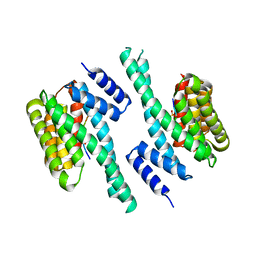 | | 14-3-3 with peptide | | Descriptor: | 14-3-3 protein zeta/delta, ARG-SER-LEU-SEP-ALA-PRO-GLY, LYS-LEU-SEP-LEU-GLN | | Authors: | Dhagat, U, Langendorf, C.G, Parker, M.W.P, Oakhill, J.S, Scott, J.W. | | Deposit date: | 2018-08-16 | | Release date: | 2019-10-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | 14-3-3 with peptide
To Be Published
|
|
6I5F
 
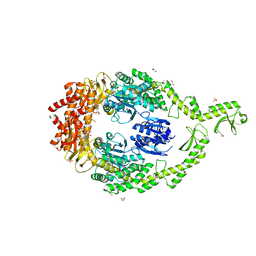 | | Crystal structure of DNA-free E.coli MutS P839E dimer mutant | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA mismatch repair protein MutS, GLYCEROL, ... | | Authors: | Bhairosing-Kok, D, Groothuizen, F.S, Fish, A, Dharadhar, S, Winterwerp, H.H.K, Sixma, T.K. | | Deposit date: | 2018-11-13 | | Release date: | 2019-08-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Sharp kinking of a coiled-coil in MutS allows DNA binding and release.
Nucleic Acids Res., 47, 2019
|
|
6EGK
 
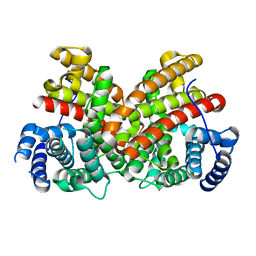 | | T181N Cucumene Synthase | | Descriptor: | Cucumene Synthase | | Authors: | Blank, P.N, Pemberton, T.A, Christianson, D.W. | | Deposit date: | 2018-08-20 | | Release date: | 2018-10-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal Structure of Cucumene Synthase, a Terpenoid Cyclase That Generates a Linear Triquinane Sesquiterpene.
Biochemistry, 57, 2018
|
|
6I23
 
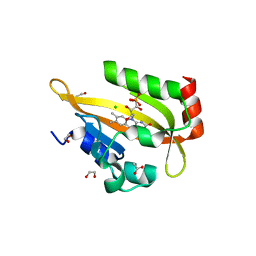 | | Flavin Analogue Sheds Light on Light-Oxygen-Voltage Domain Mechanism | | Descriptor: | 1,2-ETHANEDIOL, 1-deoxy-1-(7,8-dimethyl-2,4-dioxo-3,4-dihydropyrimido[4,5-b]quinolin-10(2H)-yl)-5-O-phosphono-D-ribitol, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Rizkallah, P.J, Kalvaitis, M.E, Allemann, R.K, Mart, R.J, Johnson, L.A. | | Deposit date: | 2018-10-31 | | Release date: | 2019-05-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Noncanonical Chromophore Reveals Structural Rearrangements of the Light-Oxygen-Voltage Domain upon Photoactivation.
Biochemistry, 58, 2019
|
|
6I6O
 
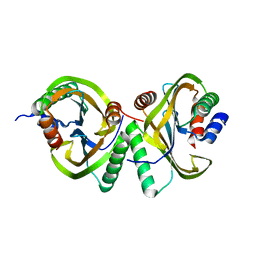 | | Circular permutant of ribosomal protein S6, swap helix 2, L75A mutant | | Descriptor: | 30S ribosomal protein S6,30S ribosomal protein S6 | | Authors: | Wang, H, Logan, D.T, Oliveberg, M. | | Deposit date: | 2018-11-15 | | Release date: | 2019-11-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Exposing the distinctive modular behavior of beta-strands and alpha-helices in folded proteins.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6E47
 
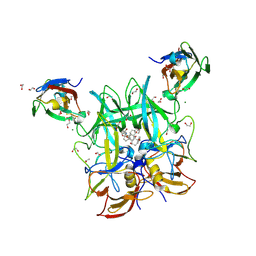 | |
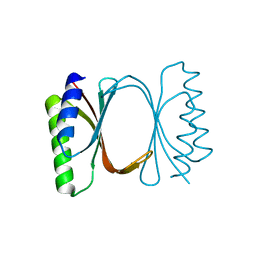
6I6Y
 
 | |
6I77
 
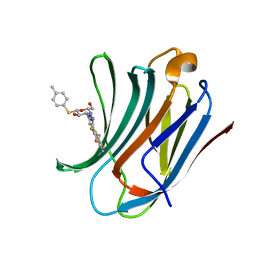 | | Galectin-3C in complex with substituted polyfluoroaryl monothiogalactoside derivative-4 | | Descriptor: | (2~{R},3~{R},4~{S},5~{R},6~{S})-4-[4-[4-azanyl-2,3,5,6-tetrakis(fluoranyl)phenyl]-1,2,3-triazol-1-yl]-2-(hydroxymethyl)-6-(4-methylphenyl)sulfanyl-oxane-3,5-diol, Galectin-3 | | Authors: | Kumar, R, Peterson, K, Nilsson, U.J, Logan, D.T. | | Deposit date: | 2018-11-15 | | Release date: | 2019-01-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.219 Å) | | Cite: | Substituted polyfluoroaryl interactions with an arginine side chain in galectin-3 are governed by steric-, desolvation and electronic conjugation effects.
Org. Biomol. Chem., 17, 2019
|
|
6I7H
 
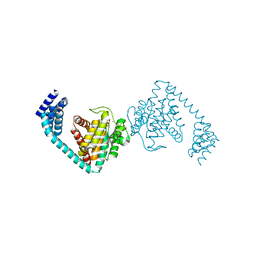 | | Crystal structure of dimeric FICD mutant K256S | | Descriptor: | Adenosine monophosphate-protein transferase FICD, MAGNESIUM ION | | Authors: | Perera, L.A, Yan, Y, Read, R.J, Ron, D. | | Deposit date: | 2018-11-16 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | An oligomeric state-dependent switch in the ER enzyme FICD regulates AMPylation and deAMPylation of BiP.
Embo J., 38, 2019
|
|
6E4F
 
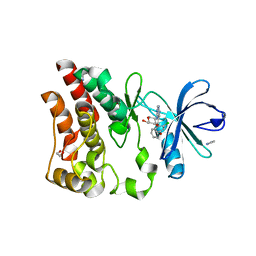 | | Crystal structure of ARQ 531 in complex with the kinase domain of BTK | | Descriptor: | 1,2-ETHANEDIOL, 1,5-anhydro-2-{[5-(2-chloro-4-phenoxybenzene-1-carbonyl)-7H-pyrrolo[2,3-d]pyrimidin-4-yl]amino}-2,3,4-trideoxy-D-erythro-hexitol, IMIDAZOLE, ... | | Authors: | Eathiraj, S. | | Deposit date: | 2018-07-17 | | Release date: | 2018-09-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | The BTK Inhibitor ARQ 531 Targets Ibrutinib-Resistant CLL and Richter Transformation.
Cancer Discov, 8, 2018
|
|
6EHS
 
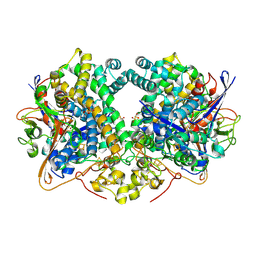 | | E. coli Hydrogenase-2 chemically reduced structure | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, DITHIONITE, FE3-S4 CLUSTER, ... | | Authors: | Carr, S.B, Evans, R.M, Beaton, S.E, Armstrong, F.A. | | Deposit date: | 2017-09-15 | | Release date: | 2018-04-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The structure of hydrogenase-2 fromEscherichia coli: implications for H2-driven proton pumping.
Biochem. J., 475, 2018
|
|
6EJC
 
 | |
6I38
 
 | |
6I8W
 
 | | Crystal structure of a membrane phospholipase A, a novel bacterial virulence factor | | Descriptor: | Alpha/beta fold hydrolase, CARBON DIOXIDE, ISOPROPYL ALCOHOL, ... | | Authors: | Granzin, J, Batra-Safferling, R. | | Deposit date: | 2018-11-21 | | Release date: | 2019-11-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural, mechanistic, and physiological insights into phospholipase A-mediated membrane phospholipid degradation in Pseudomonas aeruginosa.
Elife, 11, 2022
|
|
6I97
 
 | |
6I9V
 
 | |
6E4V
 
 | |
6EN6
 
 | | Crystal structure B of the Angiotensin-1 converting enzyme N-domain in complex with a diprolyl inhibitor. | | Descriptor: | (2~{S})-1-[(2~{S})-2-[[(1~{S})-1-[(2~{S})-1-[(2~{S})-2-azanyl-4-oxidanyl-4-oxidanylidene-butanoyl]pyrrolidin-2-yl]-2-oxidanyl-2-oxidanylidene-ethyl]amino]propanoyl]pyrrolidine-2-carboxylic acid, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Cozier, G.E, Acharya, K.R, Fienberg, S, Chibale, K, Sturrock, E.D. | | Deposit date: | 2017-10-04 | | Release date: | 2018-03-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Design and Development of a Potent and Selective Novel Diprolyl Derivative That Binds to the N-Domain of Angiotensin-I Converting Enzyme.
J. Med. Chem., 61, 2018
|
|
6E4R
 
 | | Crystal Structure of the Drosophila Melanogaster Polypeptide N-Acetylgalactosaminyl Transferase PGANT9B | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Samara, N.L, Tabak, L.A, Ten Hagen, K.G. | | Deposit date: | 2018-07-18 | | Release date: | 2018-09-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.061 Å) | | Cite: | A molecular switch orchestrates enzyme specificity and secretory granule morphology.
Nat Commun, 9, 2018
|
|
6EPB
 
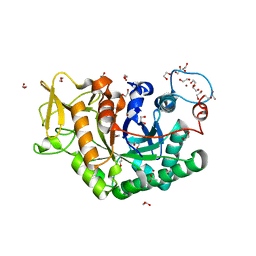 | | Structure of Chitinase 42 from Trichoderma harzianum | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Endochitinase 42, ... | | Authors: | Ramirez-Escudero, M, Jimenez-Ortega, E, Sanz-Aparicio, J. | | Deposit date: | 2017-10-11 | | Release date: | 2018-09-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Use of chitin and chitosan to produce new chitooligosaccharides by chitinase Chit42: enzymatic activity and structural basis of protein specificity.
Microb. Cell Fact., 17, 2018
|
|
