1HEM
 
 | |
1HER
 
 | |
1HEO
 
 | |
1HEP
 
 | |
1HEN
 
 | |
1HEQ
 
 | |
6HPV
 
 | | Crystal structure of mouse fetuin-B | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, Fetuin-B | | Authors: | Fahrenkamp, D, Dietzel, E, de Sanctis, D, Jovine, L. | | Deposit date: | 2018-09-22 | | Release date: | 2019-02-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of mammalian plasma fetuin-B and its mechanism of selective metallopeptidase inhibition.
Iucrj, 6, 2019
|
|
7K7R
 
 | | EBNA1 peptide AA386-405 with Fab MS39p2w174 | | Descriptor: | CHLORIDE ION, EBNA1 peptide AA386-405, Fab HC MS39p2w174, ... | | Authors: | Lanz, T.V, Robinson, W.H, Jude, K.M. | | Deposit date: | 2020-09-23 | | Release date: | 2022-01-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Clonally expanded B cells in multiple sclerosis bind EBV EBNA1 and GlialCAM.
Nature, 603, 2022
|
|
2CA6
 
 | |
6UPP
 
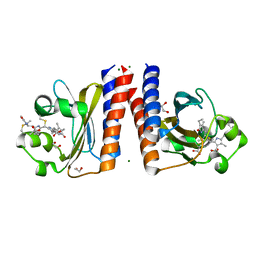 | | Radiation Damage Test of PixJ Pb state crystals | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Methyl-accepting chemotaxis protein, ... | | Authors: | Clinger, J.A, Miller, M.D, Burgie, E.S, Vierstra, R.D, Phillips Jr, G.N. | | Deposit date: | 2019-10-18 | | Release date: | 2019-12-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Photoreversible interconversion of a phytochrome photosensory module in the crystalline state.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
1NW2
 
 | | The crystal structure of the mutant R82E of Thioredoxin from Alicyclobacillus acidocaldarius | | Descriptor: | ACETATE ION, CACODYLATE ION, THIOREDOXIN, ... | | Authors: | Bartolucci, S, De Simone, G, Galdiero, S, Improta, R, Menchise, V, Pedone, C, Pedone, E, Saviano, M. | | Deposit date: | 2003-02-05 | | Release date: | 2003-08-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | An integrated structural and computational study of the thermostability of two thioredoxin mutants from Alicyclobacillus acidocaldarius
J.Bacteriol., 185, 2003
|
|
1NSW
 
 | | The Crystal Structure of the K18G Mutant of the thioredoxin from Alicyclobacillus acidocaldarius | | Descriptor: | THIOREDOXIN | | Authors: | Bartolucci, S, De Simone, G, Galdiero, S, Improta, R, Menchise, V, Pedone, C, Pedone, E, Saviano, M. | | Deposit date: | 2003-01-28 | | Release date: | 2003-08-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | An integrated structural and computational study of the thermostability of two thioredoxin mutants from Alicyclobacillus acidocaldarius
J.Bacteriol., 185, 2003
|
|
1QUW
 
 | | SOLUTION STRUCTURE OF THE THIOREDOXIN FROM BACILLUS ACIDOCALDARIUS | | Descriptor: | THIOREDOXIN | | Authors: | Nicastro, G, de Chiara, C, Pedone, E, Tato, M, Rossi, M. | | Deposit date: | 1999-07-02 | | Release date: | 2000-01-26 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of a novel thioredoxin from Bacillus acidocaldarius possible determinants of protein stability.
Eur.J.Biochem., 267, 2000
|
|
1S01
 
 | |
1VJQ
 
 | |
3VBJ
 
 | |
1AK9
 
 | | SUBTILISIN MUTANT 8321 | | Descriptor: | CALCIUM ION, ISOPROPYL ALCOHOL, SODIUM ION, ... | | Authors: | Whitlow, M, Howard, A.J, Wood, J.F. | | Deposit date: | 1997-05-30 | | Release date: | 1997-11-12 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Large increases in general stability for subtilisin BPN' through incremental changes in the free energy of unfolding.
Biochemistry, 28, 1989
|
|
1AQN
 
 | | SUBTILISIN MUTANT 8324 | | Descriptor: | CALCIUM ION, ISOPROPYL ALCOHOL, SUBTILISIN 8324, ... | | Authors: | Whitlow, M, Howard, A.J, Wood, J.F. | | Deposit date: | 1997-07-31 | | Release date: | 1998-01-14 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Large increases in general stabilityfor subtilisin BPN' through incremental changes in the free energy of unfolding
To be published
|
|
1CU2
 
 | | T4 LYSOZYME MUTANT L84M | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, LYSOZYME | | Authors: | Gassner, N.C, Baase, W.A, Lindstrom, J.D, Lu, J, Matthews, B.W. | | Deposit date: | 1999-08-20 | | Release date: | 1999-11-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Methionine and alanine substitutions show that the formation of wild-type-like structure in the carboxy-terminal domain of T4 lysozyme is a rate-limiting step in folding.
Biochemistry, 38, 1999
|
|
1CUQ
 
 | | T4 LYSOZYME MUTANT V103M | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, LYSOZYME | | Authors: | Gassner, N.C, Baase, W.A, Lindstrom, J.D, Lu, J, Matthews, B.W. | | Deposit date: | 1999-08-20 | | Release date: | 1999-11-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Methionine and alanine substitutions show that the formation of wild-type-like structure in the carboxy-terminal domain of T4 lysozyme is a rate-limiting step in folding.
Biochemistry, 38, 1999
|
|
1CV0
 
 | | T4 LYSOZYME MUTANT F104M | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, LYSOZYME | | Authors: | Gassner, N.C, Baase, W.A, Lindstrom, J.D, Lu, J, Matthews, B.W. | | Deposit date: | 1999-08-20 | | Release date: | 1999-11-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Methionine and alanine substitutions show that the formation of wild-type-like structure in the carboxy-terminal domain of T4 lysozyme is a rate-limiting step in folding.
Biochemistry, 38, 1999
|
|
1D2Y
 
 | | N-TERMINAL DOMAIN CORE METHIONINE MUTATION | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, LYSOZYME | | Authors: | Gassner, N.C, Matthews, B.W. | | Deposit date: | 1999-09-28 | | Release date: | 1999-10-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Use of differentially substituted selenomethionine proteins in X-ray structure determination.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1CV1
 
 | | T4 LYSOZYME MUTANT V111M | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, LYSOZYME | | Authors: | Gassner, N.C, Baase, W.A, Lindstrom, J.D, Lu, J, Matthews, B.W. | | Deposit date: | 1999-08-20 | | Release date: | 1999-11-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Methionine and alanine substitutions show that the formation of wild-type-like structure in the carboxy-terminal domain of T4 lysozyme is a rate-limiting step in folding.
Biochemistry, 38, 1999
|
|
1CV5
 
 | | T4 LYSOZYME MUTANT L133M | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, LYSOZYME | | Authors: | Gassner, N.C, Baase, W.A, Lindstrom, J, Lu, J, Matthews, B.W. | | Deposit date: | 1999-08-22 | | Release date: | 1999-11-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Methionine and alanine substitutions show that the formation of wild-type-like structure in the carboxy-terminal domain of T4 lysozyme is a rate-limiting step in folding.
Biochemistry, 38, 1999
|
|
1D3F
 
 | | N-TERMINAL DOMAIN CORE METHIONINE MUTATION | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, LYSOZYME | | Authors: | Gassner, N.C, Matthews, B.W. | | Deposit date: | 1999-09-29 | | Release date: | 1999-10-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Use of differentially substituted selenomethionine proteins in X-ray structure determination.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
