7OHS
 
 | |
7OHW
 
 | |
7OHV
 
 | |
7OHX
 
 | |
7OHP
 
 | |
7DLZ
 
 | | Crystal Structure of Methyltransferase Ribozyme | | Descriptor: | RNA (45-MER), U1 small nuclear ribonucleoprotein A | | Authors: | Gan, J.H, Gao, Y.Q, Jiang, H.Y, Chen, D.R, Murchie, A.I.H. | | Deposit date: | 2020-11-30 | | Release date: | 2021-10-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.002 Å) | | Cite: | The identification and characterization of a selected SAM-dependent methyltransferase ribozyme that is present in natural sequences
Nat Catal, 4, 2021
|
|
7DWH
 
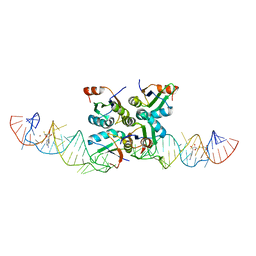 | | Complex structure of SAM-dependent methyltransferase ribozyme | | Descriptor: | COPPER (II) ION, RNA (45-MER), S-ADENOSYLMETHIONINE, ... | | Authors: | Jiang, H.Y, Gao, Y.Q, Chen, D.R, Murchie, A. | | Deposit date: | 2021-01-17 | | Release date: | 2021-10-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The identification and characterization of a selected SAM-dependent methyltransferase ribozyme that is present in natural sequences
Nat Catal, 4, 2021
|
|
7AAF
 
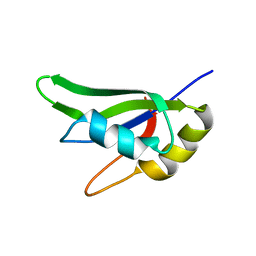 | |
7AAO
 
 | |
7OCX
 
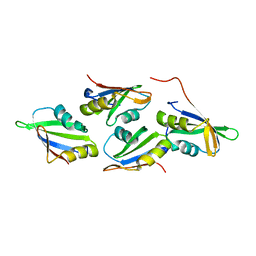 | |
7D2Y
 
 | | complex of two RRM domains | | Descriptor: | Embryonic developmental protein tofu-6, RRM2, SULFATE ION | | Authors: | Wang, X, Liao, S, Xu, C. | | Deposit date: | 2020-09-17 | | Release date: | 2021-08-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Molecular basis for PICS-mediated piRNA biogenesis and cell division.
Nat Commun, 12, 2021
|
|
7D1L
 
 | | complex structure of two RRM domains | | Descriptor: | Embryonic developmental protein tofu-6, Uncharacterized protein | | Authors: | Wang, X, Liao, S, Xu, C. | | Deposit date: | 2020-09-14 | | Release date: | 2021-08-25 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Molecular basis for PICS-mediated piRNA biogenesis and cell division.
Nat Commun, 12, 2021
|
|
7OQE
 
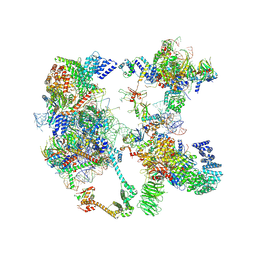 | | Saccharomyces cerevisiae spliceosomal pre-A complex (delta BS-A ACT1) | | Descriptor: | 56 kDa U1 small nuclear ribonucleoprotein component, ACT1 pre-mRNA (delta BS-A), Cold sensitive U2 snRNA suppressor 1, ... | | Authors: | Zhang, Z, Rigo, N, Dybkov, O, Fourmann, J, Will, C.L, Kumar, V, Urlaub, H, Stark, H, Luehrmann, R. | | Deposit date: | 2021-06-03 | | Release date: | 2021-08-11 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (5.9 Å) | | Cite: | Structural insights into how Prp5 proofreads the pre-mRNA branch site.
Nature, 596, 2021
|
|
7OQB
 
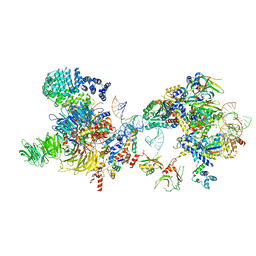 | | The U2 part of Saccharomyces cerevisiae spliceosomal pre-A complex (delta BS-A ACT1) | | Descriptor: | ACT1 pre-mRNA (delta-BS-A), Cold sensitive U2 snRNA suppressor 1, Pre-mRNA-processing ATP-dependent RNA helicase PRP5, ... | | Authors: | Zhang, Z, Rigo, N, Dybkov, O, Fourmann, J, Will, C.L, Kumar, V, Urlaub, H, Stark, H, Luehrmann, R. | | Deposit date: | 2021-06-03 | | Release date: | 2021-08-11 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Structural insights into how Prp5 proofreads the pre-mRNA branch site.
Nature, 596, 2021
|
|
7OQC
 
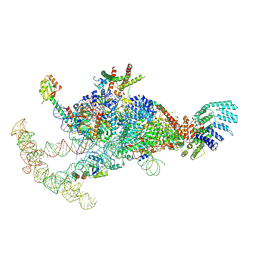 | | The U1 part of Saccharomyces cerevisiae spliceosomal pre-A complex (delta BS-A ACT1) | | Descriptor: | 56 kDa U1 small nuclear ribonucleoprotein component, ACT1 pre-mRNA (delta BS-A),ACT1 pre-mRNA (delta BS-A),ACT1 pre-mRNA (delta BS-A),ACT1 pre-mRNA (delta BS-A),ACT1 pre-mRNA (delta BS-A),ACT1 pre-mRNA (delta BS-A),ACT1 pre-mRNA (delta BS-A),ACT1 pre-mRNA (delta BS-A),ACT1 pre-mRNA (delta BS-A), Pre-mRNA-processing factor 39, ... | | Authors: | Zhang, Z, Rigo, N, Dybkov, O, Fourmann, J, Will, C.L, Kumar, V, Urlaub, H, Stark, H, Luehrmann, R. | | Deposit date: | 2021-06-03 | | Release date: | 2021-08-11 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural insights into how Prp5 proofreads the pre-mRNA branch site.
Nature, 596, 2021
|
|
7ONB
 
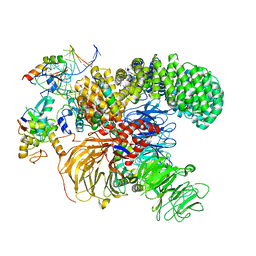 | | Structure of the U2 5' module of the A3'-SSA complex | | Descriptor: | MINX, PHD finger-like domain-containing protein 5A, RNU2, ... | | Authors: | Cretu, C, Pena, V. | | Deposit date: | 2021-05-25 | | Release date: | 2021-08-04 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of intron selection by U2 snRNP in the presence of covalent inhibitors.
Nat Commun, 12, 2021
|
|
7JLY
 
 | |
7MI2
 
 | | Seb1-G476S RNA binding domain | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Rpb7-binding protein seb1 | | Authors: | Jacewicz, A, Shuman, S. | | Deposit date: | 2021-04-16 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Genetic screen for suppression of transcriptional interference identifies a gain-of-function mutation in Pol2 termination factor Seb1.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6Z29
 
 | | Structure of eIF4G1 (37-49) - PUB1 RRM3 chimera in solution | | Descriptor: | Eukaryotic initiation factor 4F subunit p150,Nuclear and cytoplasmic polyadenylated RNA-binding protein PUB1 | | Authors: | Chaves-Arquero, B, Martinez-Lumbreras, S, Perez-Canadillas, J.M. | | Deposit date: | 2020-05-15 | | Release date: | 2021-05-26 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | eIF4G1 N-terminal intrinsically disordered domain is a multi-docking station for RNA, Pab1, Pub1, and self-assembly.
Front Mol Biosci, 9, 2022
|
|
7LMA
 
 | | Tetrahymena telomerase T3D2 structure at 3.3 Angstrom | | Descriptor: | Telomerase La-related protein p65, Telomerase RNA, Telomerase associated protein p50, ... | | Authors: | He, Y, Wang, Y, Liu, B, Helmling, C, Susac, L, Cheng, R, Zhou, Z.H, Feigon, J. | | Deposit date: | 2021-02-05 | | Release date: | 2021-05-12 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structures of telomerase at several steps of telomere repeat synthesis.
Nature, 593, 2021
|
|
7LMB
 
 | | Tetrahymena telomerase T5D5 structure at 3.8 Angstrom | | Descriptor: | Telomerase La-related protein p65, Telomerase RNA, Telomerase associated protein p50, ... | | Authors: | He, Y, Wang, Y, Liu, B, Helmling, C, Susac, L, Cheng, R, Zhou, Z.H, Feigon, J. | | Deposit date: | 2021-02-05 | | Release date: | 2021-05-12 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structures of telomerase at several steps of telomere repeat synthesis.
Nature, 593, 2021
|
|
7LRQ
 
 | | Crystal structure of human SFPQ/NONO heterodimer, conserved DBHS region | | Descriptor: | CHLORIDE ION, Non-POU domain-containing octamer-binding protein, Splicing factor, ... | | Authors: | Marshall, A.C, Bond, C.S, Mohnen, I. | | Deposit date: | 2021-02-17 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Paraspeckle subnuclear bodies depend on dynamic heterodimerisation of DBHS RNA-binding proteins via their structured domains.
J.Biol.Chem., 298, 2022
|
|
7LRU
 
 | | Crystal structure of SFPQ-NONO-SFPQ chimeric protein homodimer | | Descriptor: | Splicing factor, proline- and glutamine-rich,Isoform 2 of Non-POU domain-containing octamer-binding protein,Isoform Short of Splicing factor, proline- and glutamine-rich | | Authors: | Marshall, A.C, Bond, C.S, Mohnen, I, Knott, G.J. | | Deposit date: | 2021-02-17 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of SFPQ-NONO-SFPQ chimeric protein homodimer
To Be Published
|
|
7C36
 
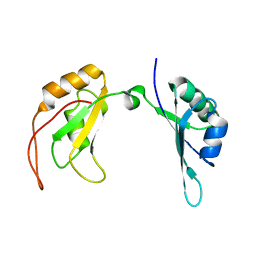 | | c-Myc DNA binding protein structure | | Descriptor: | RNA-binding motif, single-stranded-interacting protein 1 | | Authors: | Aggarwal, P, Bhavesh, N.S. | | Deposit date: | 2020-05-11 | | Release date: | 2021-05-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Hinge like domain motion facilitates human RBMS1 protein binding to proto-oncogene c-myc promoter.
Nucleic Acids Res., 49, 2021
|
|
6WMZ
 
 | | Crystal structure of human SFPQ/NONO complex | | Descriptor: | Non-POU domain-containing octamer-binding protein, SULFATE ION, Splicing factor, ... | | Authors: | Lee, M, Bond, C.S. | | Deposit date: | 2020-04-22 | | Release date: | 2021-04-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure analysis of human SFPQ/NONO complex
To Be Published
|
|
