6XVR
 
 | | Human myelin protein P2 mutant L35S | | Descriptor: | Myelin P2 protein, PALMITIC ACID, SULFATE ION | | Authors: | Ruskamo, S, Lehtimaki, M, Kursula, P. | | Deposit date: | 2020-01-22 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Cryo-EM, X-ray diffraction, and atomistic simulations reveal determinants for the formation of a supramolecular myelin-like proteolipid lattice.
J.Biol.Chem., 295, 2020
|
|
6XVE
 
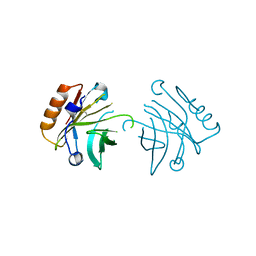 | |
5H9A
 
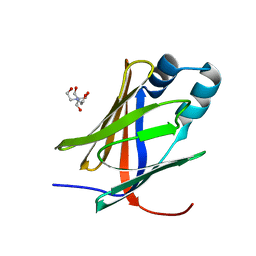 | | Crystal structure of the Apo form of human cellular retinol binding protein 1 | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Retinol-binding protein 1 | | Authors: | Golczak, M, Arne, J.M, Silvaroli, J.A, Kiser, P.D, Banerjee, S. | | Deposit date: | 2015-12-26 | | Release date: | 2016-03-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.381 Å) | | Cite: | Ligand Binding Induces Conformational Changes in Human Cellular Retinol-binding Protein 1 (CRBP1) Revealed by Atomic Resolution Crystal Structures.
J.Biol.Chem., 291, 2016
|
|
6WNJ
 
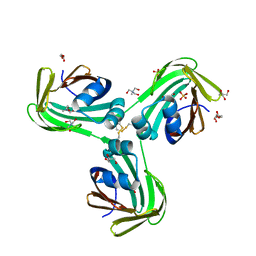 | |
4LZU
 
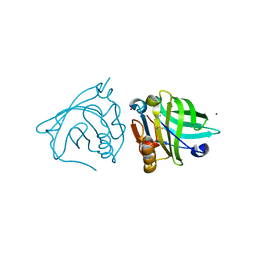 | |
5K06
 
 | | Recombinant bovine beta-lactoglobulin with uncleaved N-terminal methionine (rBlgB) | | Descriptor: | Beta-lactoglobulin, GLYCEROL, MYRISTIC ACID, ... | | Authors: | Loch, J.I, Bonarek, P, Tworzydlo, M, Polit, A, Hawro, B, Lach, A, Ludwin, E, Lewinski, K. | | Deposit date: | 2016-05-17 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Engineered beta-Lactoglobulin Produced in E. coli: Purification, Biophysical and Structural Characterisation.
Mol Biotechnol., 58, 2016
|
|
4K19
 
 | | The structure of Human Siderocalin bound to the bacterial siderophore fluvibactin | | Descriptor: | (4S,5R)-N,N-bis{3-[(2,3-dihydroxybenzoyl)amino]propyl}-2-(2,3-dihydroxyphenyl)-5-methyl-4,5-dihydro-1,3-oxazole-4-carboxamide, CHLORIDE ION, FE (III) ION, ... | | Authors: | Correnti, C, Clifton, M.C, Strong, R.K. | | Deposit date: | 2013-04-04 | | Release date: | 2013-07-31 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Siderocalin Outwits the Coordination Chemistry of Vibriobactin, a Siderophore of Vibrio cholerae.
Acs Chem.Biol., 8, 2013
|
|
4NNT
 
 | |
4NNS
 
 | |
4MVK
 
 | |
4NLJ
 
 | | Crystal structure of sheep beta-lactoglobulin (space group P1) | | Descriptor: | Beta-lactoglobulin-1/B, SULFATE ION | | Authors: | Loch, J.I, Molenda, M, Kopec, M, Swiatek, S, Lewinski, K. | | Deposit date: | 2013-11-14 | | Release date: | 2014-03-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of two crystal forms of sheep beta-lactoglobulin with EF-loop in closed conformation
Biopolymers, 101, 2014
|
|
4NLI
 
 | | Crystal structure of sheep beta-lactoglobulin (space group P3121) | | Descriptor: | Beta-lactoglobulin-1/B, SULFATE ION | | Authors: | Loch, J.I, Molenda, M, Kopec, M, Swiatek, S, Lewinski, K. | | Deposit date: | 2013-11-14 | | Release date: | 2014-03-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of two crystal forms of sheep beta-lactoglobulin with EF-loop in closed conformation
Biopolymers, 101, 2014
|
|
2K23
 
 | |
2IFB
 
 | |
2HZR
 
 | | Crystal structure of human apolipoprotein D (ApoD) | | Descriptor: | Apolipoprotein D | | Authors: | Eichinger, A, Skerra, A. | | Deposit date: | 2006-08-09 | | Release date: | 2007-08-14 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insight into the dual ligand specificity and mode of high density lipoprotein association of apolipoprotein d.
J.Biol.Chem., 282, 2007
|
|
2JN3
 
 | | NMR structure of cl-BABP complexed to chenodeoxycholic acid | | Descriptor: | CHENODEOXYCHOLIC ACID, Fatty acid-binding protein, liver | | Authors: | Eliseo, T, Ragona, L, Catalano, M, Assfalf, M, Paci, M, Zetta, L, Molinari, H, Cicero, D.O. | | Deposit date: | 2006-12-22 | | Release date: | 2007-07-03 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Structural and dynamic determinants of ligand binding in the ternary complex of chicken liver bile acid binding protein with two bile salts revealed by NMR
Biochemistry, 46, 2007
|
|
2K62
 
 | | NMR solution structure of the supramolecular adduct between a liver cytosolic bile acid binding protein and a bile acid-based Gd(III)-chelate | | Descriptor: | (3alpha,5alpha,8alpha)-3-[(N,N-bis{2-[bis(carboxymethyl)amino]ethyl}-L-gamma-glutamyl)amino]cholan-24-oic acid, Liver fatty acid-binding protein, YTTERBIUM (III) ION | | Authors: | Tomaselli, S, Zanzoni, S, Ragona, L, Gianolio, E, Aime, S, Assfalg, M, Molinari, H. | | Deposit date: | 2008-07-03 | | Release date: | 2008-11-04 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the supramolecular adduct between a liver cytosolic bile acid binding protein and a bile acid-based gadolinium(III)-chelate, a potential hepatospecific magnetic resonance imaging contrast agent.
J.Med.Chem., 51, 2008
|
|
2HZQ
 
 | |
2HNX
 
 | | Crystal Structure of aP2 | | Descriptor: | ACETIC ACID, Fatty acid-binding protein, adipocyte, ... | | Authors: | Marr, E, Tardie, M, Carty, M, Brown Phillips, T, Qiu, X, Karam, G. | | Deposit date: | 2006-07-13 | | Release date: | 2006-11-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Expression, purification, crystallization and structure of human adipocyte lipid-binding protein (aP2).
Acta Crystallogr.,Sect.F, 62, 2006
|
|
2KT4
 
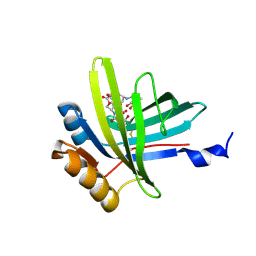 | | Lipocalin Q83 is a Siderocalin | | Descriptor: | Extracellular fatty acid-binding protein, GALLIUM (III) ION, N,N',N''-[(3S,7S,11S)-2,6,10-trioxo-1,5,9-trioxacyclododecane-3,7,11-triyl]tris(2,3-dihydroxybenzamide) | | Authors: | Coudevylle, N, Geist, L, Hartl, M, Kontaxis, G, Bister, K, Konrat, R. | | Deposit date: | 2010-01-18 | | Release date: | 2010-09-08 | | Last modified: | 2016-01-27 | | Method: | SOLUTION NMR | | Cite: | The v-myc-induced Q83 lipocalin is a siderocalin.
J.Biol.Chem., 285, 2010
|
|
2KTD
 
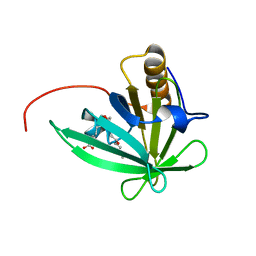 | | Solution structure of mouse lipocalin-type prostaglandin D synthase / substrate analog (U-46619) complex | | Descriptor: | (5Z)-7-{(1R,4S,5S,6R)-6-[(1E,3S)-3-hydroxyoct-1-en-1-yl]-2-oxabicyclo[2.2.1]hept-5-yl}hept-5-enoic acid, Prostaglandin-H2 D-isomerase | | Authors: | Shimamoto, S, Maruo, H, Yoshida, T, Kato, N, Ohkubo, T. | | Deposit date: | 2010-01-27 | | Release date: | 2011-02-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Lipocalin-type Prostaglandin D synthase / Substrate analog complex reveals Open-Closed Conformational Change required for Substrate Recognition
To be Published
|
|
2GJ5
 
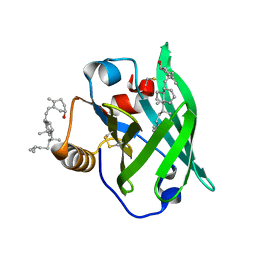 | | Crystal structure of a secondary vitamin D3 binding site of milk beta-lactoglobulin | | Descriptor: | (1S,3Z)-3-[(2E)-2-[(1R,3AR,7AS)-7A-METHYL-1-[(2R)-6-METHYLHEPTAN-2-YL]-2,3,3A,5,6,7-HEXAHYDRO-1H-INDEN-4-YLIDENE]ETHYLI DENE]-4-METHYLIDENE-CYCLOHEXAN-1-OL, Beta-lactoglobulin | | Authors: | Yang, M.C, Guan, H.H, Liu, M.Y, Yang, J.M, Chen, W.L, Chen, C.J, Mao, S.J. | | Deposit date: | 2006-03-30 | | Release date: | 2007-10-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of a secondary vitamin D3 binding site of milk beta-lactoglobulin.
Proteins, 71, 2008
|
|
2HMB
 
 | | THREE-DIMENSIONAL STRUCTURE OF RECOMBINANT HUMAN MUSCLE FATTY ACID-BINDING PROTEIN | | Descriptor: | MUSCLE FATTY ACID BINDING PROTEIN, PALMITIC ACID | | Authors: | Zanotti, G, Scapin, G, Spadon, P, Veerkamp, J.H, Sacchettini, J.C. | | Deposit date: | 1992-09-11 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Three-dimensional structure of recombinant human muscle fatty acid-binding protein.
J.Biol.Chem., 267, 1992
|
|
2LB6
 
 | | Structure of 18694Da MUP, typical to the major urinary protein family: MUP9, MUP11, MUP15, MUP18 & MUP19 | | Descriptor: | Major urinary protein 6 | | Authors: | Phelan, M.M, Mclean, L, Beynon, R.J, Hurst, J.L, Lian, L. | | Deposit date: | 2011-03-23 | | Release date: | 2012-03-28 | | Last modified: | 2023-12-06 | | Method: | SOLUTION NMR | | Cite: | Structural insights into the specificity of darcin, an atypical major urinary protein.
To be Published
|
|
2HLV
 
 | | Bovine Odorant Binding Protein deswapped triple mutant | | Descriptor: | 3,6-BIS(METHYLENE)DECANOIC ACID, GLYCEROL, Odorant-binding protein | | Authors: | Spinelli, S, Tegoni, M, Cambillau, C. | | Deposit date: | 2006-07-10 | | Release date: | 2008-03-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Deswapping bovine odorant binding protein.
Biochim.Biophys.Acta, 1784, 2008
|
|
