9EQ5
 
 | |
9ENF
 
 | |
9C0J
 
 | |
9F3Z
 
 | |
9G13
 
 | | VHH H3-2 in complex with Tau C-terminal peptide | | Descriptor: | Isoform Tau-F of Microtubule-associated protein tau, VHH H3-2 | | Authors: | Dupre, E, Landrieu, I, Danis, C, Hanoulle, X, Mortelecque, J. | | Deposit date: | 2024-07-09 | | Release date: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | VHH H3-2 in complex with Tau C-terminal peptide
To Be Published
|
|
9F7W
 
 | |
9EO6
 
 | | SARS-CoV2 major protease in complex with a covalent inhibitor SLL11. | | Descriptor: | 3C-like proteinase nsp5, Inhibitor SLL11, POTASSIUM ION | | Authors: | Moche, M, Lennerstrand, J, Nyman, T, Strandback, E, Akaberi, D. | | Deposit date: | 2024-03-14 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Identification of novel and potent inhibitors of SARS-CoV-2 main protease from DNA-encoded chemical libraries.
Antimicrob.Agents Chemother., 68, 2024
|
|
9G2A
 
 | | Staphylococcus aureus MazF in complex with nanobody 4 | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Endoribonuclease MazF, ... | | Authors: | Prolic-Kalinsek, M, Zorzini, V, Haesaerts, S, Loris, R. | | Deposit date: | 2024-07-10 | | Release date: | 2024-07-17 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2.046594 Å) | | Cite: | Staphylococcus aureus MazF in complex with nanobody 4.
To Be Published
|
|
9BDX
 
 | | NF-kappaB RelA homo-dimer bound to CG-centric kappaB DNA | | Descriptor: | DNA (5'-D(*AP*TP*CP*AP*CP*TP*GP*GP*AP*AP*GP*TP*TP*CP*CP*CP*AP*GP*T)-3'), DNA (5'-D(P*AP*CP*TP*GP*GP*GP*AP*AP*CP*TP*TP*CP*CP*AP*GP*TP*GP*AP*T)-3'), Transcription factor p65 | | Authors: | Biswas, T, Shahabi, S, Tsodikov, O.V, Ghosh, G. | | Deposit date: | 2024-04-12 | | Release date: | 2024-04-24 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Transient interactions modulate the affinity of NF-kappa B transcription factors for DNA.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
9C7U
 
 | | Structure of the human truncated BOS complex in GDN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BOS complex subunit NOMO2, Nicalin, ... | | Authors: | Nguyen, V.N, Tomaleri, G.P, Voorhees, R.M. | | Deposit date: | 2024-06-11 | | Release date: | 2024-08-21 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | Role of a holo-insertase complex in the biogenesis of biophysically diverse ER membrane proteins.
Mol.Cell, 84, 2024
|
|
9BF6
 
 | |
9C9R
 
 | |
9DK4
 
 | | ancestral hydroxynitrile lyase with fifteen substitutions | | Descriptor: | Fifteen-substitution variant of an ancestral hydroxynitrile lyase, SODIUM ION | | Authors: | Sarak, S.C, Tan, P, Evans III, R.L, Shi, K, Kazlauskas, R.J, Aihara, H, Pierce, C.T, Walsh, M.E, Greenberg, L.R. | | Deposit date: | 2024-09-07 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | To be published
To Be Published
|
|
6WTS
 
 | | CryoEM structure of the C. sordellii lethal toxin TcsL in complex with SEMA6A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, SEMA6A, ... | | Authors: | Kucharska, I, Rubinstein, J.L, Julien, J.P. | | Deposit date: | 2020-05-03 | | Release date: | 2020-07-08 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Recognition of Semaphorin Proteins by P. sordellii Lethal Toxin Reveals Principles of Receptor Specificity in Clostridial Toxins.
Cell, 182, 2020
|
|
9CYS
 
 | | Toxin/immunity complex for a T6SS lipase effector from E. cloacae | | Descriptor: | Ankyrin repeat domain-containing protein, CITRIC ACID, GLYCEROL, ... | | Authors: | Cuthbert, B.J, Jensen, S.J, Goulding, C.W, Hayes, C.S. | | Deposit date: | 2024-08-02 | | Release date: | 2024-08-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Advanced glycation end-product crosslinking activates a type VI secretion system phospholipase effector protein.
Nat Commun, 15, 2024
|
|
9FYO
 
 | | Lacto-N-biosidase from Trueperella pyogenes | | Descriptor: | NICKEL (II) ION, TrpyGH20, beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Vuillemin, M, Siebenhaar, S, Zeuner, B, Morth, J.P. | | Deposit date: | 2024-07-03 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Discovery of Lacto-N-biosidases and a Novel N-Acetyllactosaminidase Activity in the CAZy Family GH20: Functional Diversity and Structural Insights.
Chembiochem, 2024
|
|
9DNA
 
 | |
9CJ8
 
 | | Lineage IV Lassa virus glycoprotein (Josiah) in complex with rabbit polyclonal antibody (LAVA01-like epitope) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Brouwer, P.J.M, Perrett, H.R, Ward, A.B. | | Deposit date: | 2024-07-05 | | Release date: | 2024-09-25 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.74 Å) | | Cite: | Defining bottlenecks and opportunities for Lassa virus neutralization by structural profiling of vaccine-induced polyclonal antibody responses.
Cell Rep, 43, 2024
|
|
9F44
 
 | |
9G7K
 
 | |
9C4O
 
 | |
9EOX
 
 | | SARS-CoV2 major protease in covalent complex with a soluble inhibitor. | | Descriptor: | 3C-like proteinase nsp5, POTASSIUM ION, Soluble inhibitor | | Authors: | Moche, M, Lennerstrand, J, Nyman, T, Strandback, E, Akaberi, D. | | Deposit date: | 2024-03-15 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Identification of novel and potent inhibitors of SARS-CoV-2 main protease from DNA-encoded chemical libraries.
Antimicrob.Agents Chemother., 68, 2024
|
|
9GAC
 
 | |
3BYN
 
 | |
9BDL
 
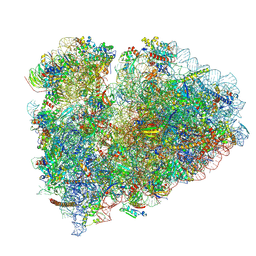 | | 80S ribosome with angiogenin | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S10, ... | | Authors: | Loveland, A.B, Korostelev, A.A. | | Deposit date: | 2024-04-12 | | Release date: | 2024-07-24 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural mechanism of angiogenin activation by the ribosome.
Nature, 630, 2024
|
|
