7B9Y
 
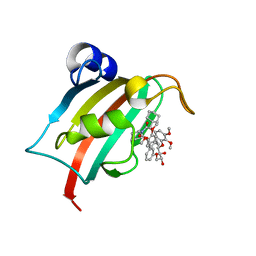 | | Structure of the FKBP51FK1 domain in complex with the macrocyclic SAFit analogue 64a | | Descriptor: | 2-cyclohexyl-12-[2-(3,4-dimethoxyphenyl)ethyl]-20,21-dihydroxy-25,26-dimethoxy-11,18,23-trioxa-4-azatetracyclo[22.3.1.113,17.04,9]nonacosa-1(27),13(29),14,16,24(28),25-hexaene-3,10-dione, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Bauder, M, Meyners, C, Purder, P, Merz, S, Voll, A, Heymann, T, Hausch, F. | | Deposit date: | 2020-12-15 | | Release date: | 2021-03-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure-Based Design of High-Affinity Macrocyclic FKBP51 Inhibitors.
J.Med.Chem., 64, 2021
|
|
8S0M
 
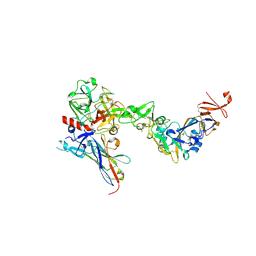 | | Crystal structure of the HKU1 receptor binding domain in complex with TMPRSS2 and the nanobody A01 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody A01, ... | | Authors: | Duquerroy, S, Fernandez, I, Rey, F. | | Deposit date: | 2024-02-14 | | Release date: | 2024-06-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.55 Å) | | Cite: | Structural basis of TMPRSS2 zymogen activation and recognition by the HKU1 seasonal coronavirus.
Cell, 187, 2024
|
|
8B3E
 
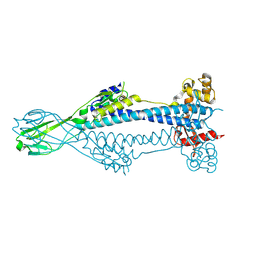 | | Variant Surface Glycoprotein VSG397 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Variant surface glycoprotein 397 | | Authors: | Zeelen, J.P, Stebbins, C.E, Dakovic, S, Foti, K. | | Deposit date: | 2022-09-16 | | Release date: | 2023-01-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Structural similarities between the metacyclic and bloodstream form variant surface glycoproteins of the African trypanosome.
Plos Negl Trop Dis, 17, 2023
|
|
8RPJ
 
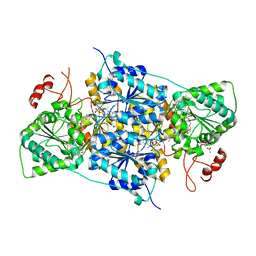 | | JanthE from Janthinobacterium sp. HH01 | | Descriptor: | ACETATE ION, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Lanza, L, Leogrande, C, Rabe von Pappenheim, F, Tittmann, K, Mueller, M. | | Deposit date: | 2024-01-16 | | Release date: | 2024-06-12 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification and Characterization of Thiamine Diphosphate-Dependent Lyases with an Unusual CDG Motif.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
7S57
 
 | | Structure of Sortase A from Streptococcus pyogenes with the b7-b8 loop sequence of Enterococcus faecalis Sortase A | | Descriptor: | Class A sortase, sortase A chimera | | Authors: | Svendsen, J.E, Johnson, D.A, Gao, M, Antos, J.M, Amacher, J.F. | | Deposit date: | 2021-09-09 | | Release date: | 2022-01-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and biochemical analyses of selectivity determinants in chimeric Streptococcus Class A sortase enzymes.
Protein Sci., 31, 2022
|
|
8BE1
 
 | | SARS-CoV-2 RBD in complex with a Fab fragment of a neutralising antibody mRBD2 | | Descriptor: | Antibody heavy chain, Antibody light chain, SULFATE ION, ... | | Authors: | Lulla, A, Brear, P, Fischer, K, Hollfelder, F, Hyvonen, M. | | Deposit date: | 2022-10-21 | | Release date: | 2023-01-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Microfluidics-enabled fluorescence-activated cell sorting of single pathogen-specific antibody secreting cells for the rapid discovery of monoclonal antibodies
Biorxiv, 2023
|
|
6DV9
 
 | | Crystal structure of Mycobacterium tuberculosis transcription initiation complex(ECF sigma factor L) containing 5nt RNA with 4nt spacer | | Descriptor: | DNA (5'-D(*GP*CP*AP*TP*CP*CP*GP*TP*GP*AP*GP*TP*CP*GP*AP*GP*G)-3'), DNA (5'-D(P*CP*GP*TP*GP*TP*CP*AP*GP*AP*GP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*GP*C)-3'), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Lin, W, Das, K, Feng, Y, Ebright, R.H. | | Deposit date: | 2018-06-23 | | Release date: | 2019-02-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structural basis of ECF-sigma-factor-dependent transcription initiation.
Nat Commun, 10, 2019
|
|
7S53
 
 | | Structure of Sortase A from Streptococcus pyogenes with the b7-b8 loop sequence from Listeria monocytogenes Sortase A | | Descriptor: | Class A sortase, sortase A chimera | | Authors: | Johnson, D.A, Svendsen, J.E, Antos, J.M, Amacher, J.F. | | Deposit date: | 2021-09-09 | | Release date: | 2022-01-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and biochemical analyses of selectivity determinants in chimeric Streptococcus Class A sortase enzymes.
Protein Sci., 31, 2022
|
|
7N3I
 
 | | Crystal structure of the SARS-CoV-2 receptor binding domain in complex with the human neutralizing antibody Fab fragment C098 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, C098 Fab heavy chain, C098 Fab light chain, ... | | Authors: | Flyak, A.I, Bjorkman, P.J, Barnes, C.O. | | Deposit date: | 2021-06-01 | | Release date: | 2021-08-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Affinity maturation of SARS-CoV-2 neutralizing antibodies confers potency, breadth, and resilience to viral escape mutations.
Immunity, 54, 2021
|
|
6DVS
 
 | | Crystal structure of Pseudomonas stutzeri D-phenylglycine aminotransferase | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, ... | | Authors: | Couture, J.F, Chica, R. | | Deposit date: | 2018-06-25 | | Release date: | 2018-09-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.821 Å) | | Cite: | Structural Determinants of the Stereoinverting Activity of Pseudomonas stutzeri d-Phenylglycine Aminotransferase.
Biochemistry, 57, 2018
|
|
7BAF
 
 | | Crystal structure of PAFB in complex with zinc | | Descriptor: | ACETATE ION, Antifungal protein, ZINC ION | | Authors: | Guagnini, F, Huber, A, Alex, J.M, Marx, F, Crowley, P.B. | | Deposit date: | 2020-12-15 | | Release date: | 2021-03-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.123 Å) | | Cite: | Porous assembly of an antifungal protein mediated by zinc and sulfonato-calix[8]arene.
J.Struct.Biol., 213, 2021
|
|
8RFD
 
 | | Crystal structure of N-terminal SARS-CoV-2 nsp1 in complex with fragment hit 7H2_AL2 refined against the anomalous diffraction data | | Descriptor: | (1~{R})-1-(4-iodophenyl)ethanamine, Host translation inhibitor nsp1 | | Authors: | Ma, S, Damfo, S, Mykhaylyk, V, Kozielski, F. | | Deposit date: | 2023-12-12 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | High-confidence placement of low-occupancy fragments into electron density using the anomalous signal of sulfur and halogen atoms.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
8RGK
 
 | | Structure of Human Serum Albumin in complex with Aristolochic Acid at 1.9 A resolution | | Descriptor: | 1,2-ETHANEDIOL, 8-methoxy-6-nitro-naphtho[1,2-e][1,3]benzodioxole-5-carboxylic acid, MYRISTIC ACID, ... | | Authors: | Pomyalov, S, Sidorenko, V.S, Grollman, A.P, Shoham, G. | | Deposit date: | 2023-12-13 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and mechanistic insights into the transport of aristolochic acids and their active metabolites by human serum albumin.
J.Biol.Chem., 300, 2024
|
|
7BAE
 
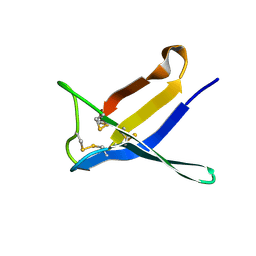 | | Crystal structure of PAFB | | Descriptor: | Antifungal protein | | Authors: | Guagnini, F, Huber, A, Alex, J.M, Marx, F, Crowley, P.B. | | Deposit date: | 2020-12-15 | | Release date: | 2021-03-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Porous assembly of an antifungal protein mediated by zinc and sulfonato-calix[8]arene.
J.Struct.Biol., 213, 2021
|
|
7N3F
 
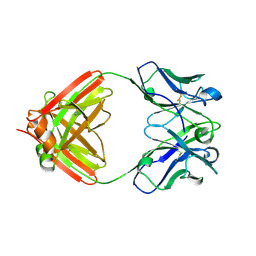 | | Crystal structure of an anti-SARS-CoV-2 human neutralizing antibody Fab fragment C080 | | Descriptor: | C080 Fab Heavy Chain, C080 Fab Light Chain | | Authors: | Flyak, A.I, Bjorkman, P.J, Barnes, C.O. | | Deposit date: | 2021-06-01 | | Release date: | 2021-08-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Affinity maturation of SARS-CoV-2 neutralizing antibodies confers potency, breadth, and resilience to viral escape mutations.
Immunity, 54, 2021
|
|
6C1V
 
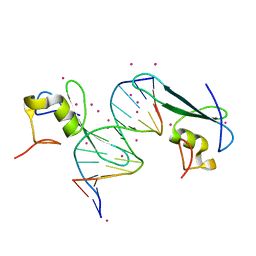 | | MBD2 in complex with double-stranded DNA | | Descriptor: | 12-mer DNA, Methyl-CpG-binding domain protein 2, UNKNOWN ATOM OR ION | | Authors: | Lei, M, Tempel, W, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-01-05 | | Release date: | 2018-02-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for the ability of MBD domains to bind methyl-CG and TG sites in DNA.
J. Biol. Chem., 293, 2018
|
|
7C20
 
 | |
7N0Z
 
 | |
7BA0
 
 | | Structure of the FKBP51FK1 domain in complex with the macrocyclic SAFit analogue 63 | | Descriptor: | 2-cyclohexyl-12-[2-(3,4-dimethoxyphenyl)ethyl]-20,21-dihydroxy-25,28-dimethoxy-11,18,23-trioxa-4-azatetracyclo[22.2.2.113,17.04,9]nonacosa-1(26),13(29),14,16,24,27-hexaene-3,10-dione, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Bauder, M, Meyners, C, Purder, P, Merz, S, Voll, A, Heymann, T, Hausch, F. | | Deposit date: | 2020-12-15 | | Release date: | 2021-03-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Structure-Based Design of High-Affinity Macrocyclic FKBP51 Inhibitors.
J.Med.Chem., 64, 2021
|
|
6C2F
 
 | | MBD2 in complex with methylated DNA | | Descriptor: | 12-mer DNA, Methyl-CpG-binding domain protein 2, UNKNOWN ATOM OR ION | | Authors: | Liu, K, Xu, C, Tempel, W, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Min, J, Structural Genomics Consortium, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-01-08 | | Release date: | 2018-02-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | MBD2 in complex with methylated DNA
to be published
|
|
7BB4
 
 | | Crystal structure of perdeuterated PLL lectin in complex with L-fucose | | Descriptor: | GLYCEROL, PLL lectin, alpha-L-fucopyranose, ... | | Authors: | Gajdos, L, Blakeley, M.P, Kumar, A, Wimmerova, M, Haertlein, M, Forsyth, V.T, Imberty, A, Devos, J.M. | | Deposit date: | 2020-12-16 | | Release date: | 2021-03-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Visualization of hydrogen atoms in a perdeuterated lectin-fucose complex reveals key details of protein-carbohydrate interactions.
Structure, 29, 2021
|
|
8RUK
 
 | | Structure of Oceanobacillus iheyensis group II intron in the presence of Na+, Mg2+, and ARN25850 | | Descriptor: | 2-[2,6-bis(bromanyl)-3,4,5-tris(oxidanyl)phenyl]carbonyl-~{N}-(2-pyrrolidin-1-ylethyl)-1-benzofuran-5-carboxamide, Domains 1-5, MAGNESIUM ION, ... | | Authors: | Silvestri, I, Marcia, M. | | Deposit date: | 2024-01-31 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (4.81 Å) | | Cite: | Targeting the conserved active site of splicing machines with specific and selective small molecule modulators.
Nat Commun, 15, 2024
|
|
7NAA
 
 | | Crystal structure of Mycobacterium tuberculosis H37Rv PknF kinase domain | | Descriptor: | (4-{[4-(1-benzothiophen-2-yl)pyrimidin-2-yl]amino}phenyl)[4-(pyrrolidin-1-yl)piperidin-1-yl]methanone, Non-specific serine/threonine protein kinase | | Authors: | Oliveira, A.A, Cabarca, S, dos Reis, C.V, Takarada, J.E, Counago, R.M, Balan, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-06-21 | | Release date: | 2021-08-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure of the Mycobacterium tuberculosis c PknF and conformational changes induced in forkhead-associated regulatory domains.
Curr Res Struct Biol, 3, 2021
|
|
8RF6
 
 | | Crystal structure of N-terminal SARS-CoV-2 nsp1 in complex with fragment hit 11A7_AL5 refined against the anomalous diffraction data | | Descriptor: | 6-iodanyl-2,3-dihydro-1,3-benzothiazol-2-amine, Host translation inhibitor nsp1 | | Authors: | Ma, S, Damfo, S, Mykhaylyk, V, Kozielski, F. | | Deposit date: | 2023-12-12 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | High-confidence placement of low-occupancy fragments into electron density using the anomalous signal of sulfur and halogen atoms.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
7RXS
 
 | | Crystal of BRD4(D1) with 2-[(3S)-3-{5-[2-(3,5-dimethylphenoxy)pyrimidin-4-yl]-4-(4-iodophenyl)-1H-imidazol-1-yl}pyrrolidin-1-yl]ethan-1-amine | | Descriptor: | 1,2-ETHANEDIOL, 2-[(3S)-3-{5-[2-(3,5-dimethylphenoxy)pyrimidin-4-yl]-4-(4-iodophenyl)-1H-imidazol-1-yl}pyrrolidin-1-yl]ethan-1-amine, Bromodomain-containing protein 4 | | Authors: | Cui, H, Johnson, J.A, Shi, K, Aihara, H, Pomerantz, W.C.K. | | Deposit date: | 2021-08-23 | | Release date: | 2022-01-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | A Structure-based Design Approach for Generating High Affinity BRD4 D1-Selective Chemical Probes.
J.Med.Chem., 65, 2022
|
|
