1AV4
 
 | |
1OBV
 
 | | Y94F flavodoxin from Anabaena | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVODOXIN, SULFATE ION | | Authors: | Romero, A, Ramon, A, Fernandez-Cabrera, C, Irun, M.P, Sancho, J. | | Deposit date: | 2003-01-31 | | Release date: | 2003-04-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | How Fmn Binds to Anabaena Apoflavodoxin: A Hydrophobic Encounter at an Open Binding Site
J.Biol.Chem., 278, 2003
|
|
1SF7
 
 | | BINDING OF TETRA-N-ACETYLCHITOTETRAOSE TO HEW LYSOZYME: A POWDER DIFFRACTION STUDY | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LYSOZYME | | Authors: | Von Dreele, R.B. | | Deposit date: | 2004-02-19 | | Release date: | 2004-03-02 | | Last modified: | 2024-10-16 | | Method: | POWDER DIFFRACTION | | Cite: | Binding of N-acetylglucosamine oligosaccharides to hen egg-white lysozyme: a powder diffraction study.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1B3X
 
 | | XYLANASE FROM PENICILLIUM SIMPLICISSIMUM, COMPLEX WITH XYLOTRIOSE | | Descriptor: | PROTEIN (XYLANASE), beta-D-xylopyranose, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose | | Authors: | Schmidt, A, Kratky, C. | | Deposit date: | 1998-12-15 | | Release date: | 1999-04-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Xylan binding subsite mapping in the xylanase from Penicillium simplicissimum using xylooligosaccharides as cryo-protectant.
Biochemistry, 38, 1999
|
|
1O6E
 
 | | Epstein-Barr virus protease | | Descriptor: | CAPSID PROTEIN P40, PHOSPHORYLISOPROPANE | | Authors: | Buisson, M, Hernandez, J, Lascoux, D, Schoehn, G, Forest, E, Arlaud, G, Seigneurin, J, Ruigrok, R.W.H, Burmeister, W.P. | | Deposit date: | 2002-09-13 | | Release date: | 2002-11-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Crystal Structure of the Epstein-Barr Virus Protease Shows Rearrangement of the Processed C Terminus
J.Mol.Biol., 324, 2002
|
|
4ITY
 
 | | Crystal structure of Leishmania mexicana arginase | | Descriptor: | Arginase, GLYCEROL, MANGANESE (II) ION | | Authors: | D'Antonio, E.L, Ullman, B, Roberts, S.C, Gaur Dixit, U, Wilson, M.E, Hai, Y, Christianson, D.W. | | Deposit date: | 2013-01-19 | | Release date: | 2013-01-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of arginase from Leishmania mexicana and implications for the inhibition of polyamine biosynthesis in parasitic infections.
Arch.Biochem.Biophys., 535, 2013
|
|
5A79
 
 | | Novel inter-subunit contacts in Barley Stripe Mosaic Virus revealed by cryo-EM | | Descriptor: | CAPSID PROTEIN, RNA | | Authors: | Clare, D.K, Pechnikova, E, Skurat, E, Makarov, V, Sokolova, O.S, Solovyev, A.G, V Orlova, E. | | Deposit date: | 2015-07-03 | | Release date: | 2015-09-02 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Novel Inter-Subunit Contacts in Barley Stripe Mosaic Virus Revealed by Cryo-Electron Microscopy.
Structure, 23, 2015
|
|
6DDE
 
 | | Mu Opioid Receptor-Gi Protein Complex | | Descriptor: | DAMGO, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Koehl, A, Hu, H, Maeda, S, Manglik, A, Zhang, Y, Kobilka, B.K, Skiniotis, G, Weis, W.I. | | Deposit date: | 2018-05-10 | | Release date: | 2018-06-13 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of the mu-opioid receptor-Giprotein complex.
Nature, 558, 2018
|
|
3ERE
 
 | | Crystal structure of the arginine repressor protein from Mycobacterium tuberculosis in complex with the DNA operator | | Descriptor: | 5'-D(*DTP*DTP*DGP*DCP*DAP*DTP*DAP*DAP*DCP*DGP*DAP*DTP*DGP*DCP*DAP*DA)-3', 5'-D(*DTP*DTP*DGP*DCP*DAP*DTP*DCP*DGP*DTP*DTP*DAP*DTP*DGP*DCP*DAP*DA)-3', Arginine repressor, ... | | Authors: | Cherney, L.T, Cherney, M.M, Garen, C.R, Lu, G.J, James, M.N, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2008-10-01 | | Release date: | 2008-10-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the arginine repressor protein in complex with the DNA operator from Mycobacterium tuberculosis.
J.Mol.Biol., 384, 2008
|
|
4IWH
 
 | |
1AXK
 
 | | ENGINEERED BACILLUS BIFUNCTIONAL ENZYME GLUXYN-1 | | Descriptor: | CALCIUM ION, GLUXYN-1 | | Authors: | Ay, J, Heinemann, U. | | Deposit date: | 1997-10-16 | | Release date: | 1999-05-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and function of the Bacillus hybrid enzyme GluXyn-1: native-like jellyroll fold preserved after insertion of autonomous globular domain.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
3P3X
 
 | | Crystal Structure of the Cytochrome P450 Monooxygenase AurH (nterm-AurH-I) from Streptomyces Thioluteus | | Descriptor: | CHLORIDE ION, Cytochrome P450, GLYCEROL, ... | | Authors: | Zocher, G, Richter, M.E.A, Mueller, U, Hertweck, C. | | Deposit date: | 2010-10-05 | | Release date: | 2011-02-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural fine-tuning of a multifunctional cytochrome p450 monooxygenase.
J.Am.Chem.Soc., 133, 2011
|
|
3P4Q
 
 | | Crystal structure of Menaquinol:oxidoreductase in complex with oxaloacetate | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Tomasiak, T.M, Archuleta, T.L, Andrell, J, Luna-Chavez, C, Davis, T.A, Sarwar, M, Ham, A.J, McDonald, W.H, Yankowskaya, V, Stern, H.A, Johnston, J.N, Maklashina, E, Cecchini, G, Iverson, T.M. | | Deposit date: | 2010-10-06 | | Release date: | 2010-12-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Geometric restraint drives on- and off-pathway catalysis by the Escherichia coli menaquinol:fumarate reductase.
J.Biol.Chem., 286, 2011
|
|
7BL4
 
 | | in vitro reconstituted 50S-ObgE-GMPPNP-RsfS particle | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Hilal, T, Nikolay, R, Spahn, C.M.T. | | Deposit date: | 2021-01-18 | | Release date: | 2021-05-12 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Snapshots of native pre-50S ribosomes reveal a biogenesis factor network and evolutionary specialization.
Mol.Cell, 81, 2021
|
|
1V6V
 
 | | Crystal Structure Of Xylanase From Streptomyces Olivaceoviridis E-86 Complexed With 3(2)-alpha-L-arabinofuranosyl-xylotriose | | Descriptor: | ENDO-1,4-BETA-D-XYLANASE, alpha-L-arabinofuranose-(1-3)-[beta-D-xylopyranose-(1-4)]beta-D-xylopyranose-(1-4)-beta-D-xylopyranose, beta-D-xylopyranose, ... | | Authors: | Fujimoto, Z, Kaneko, S, Kuno, A, Kobayashi, H, Kusakabe, I, Mizuno, H. | | Deposit date: | 2003-12-04 | | Release date: | 2004-04-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of decorated xylooligosaccharides bound to a family 10 xylanase from Streptomyces olivaceoviridis E-86
J.Biol.Chem., 279, 2004
|
|
3TF3
 
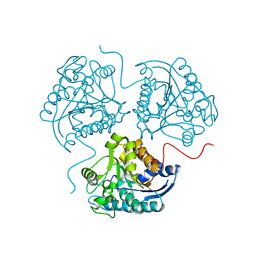 | |
3TCY
 
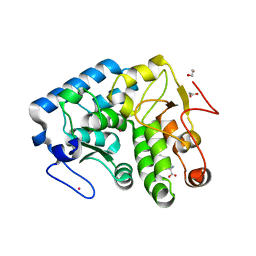 | | Crystallographic structure of phenylalanine hydroxylase from Chromobacterium violaceum (cPAH) bound to phenylalanine in a site distal to the active site | | Descriptor: | 1,2-ETHANEDIOL, COBALT (II) ION, PHENYLALANINE, ... | | Authors: | Ronau, J.A, Abu-Omar, M.M, Das, C. | | Deposit date: | 2011-08-09 | | Release date: | 2012-08-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | An additional substrate binding site in a bacterial phenylalanine hydroxylase.
Eur.Biophys.J., 42, 2013
|
|
3P6R
 
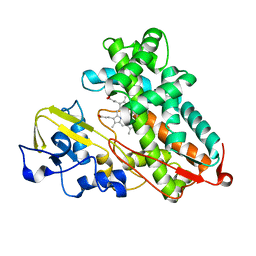 | |
8ZGT
 
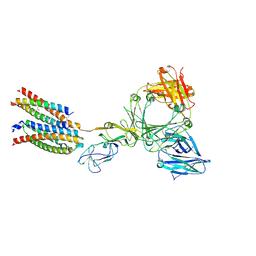 | | Structure of the ige-fc bound to its high affinity receptor fc(epsilon)ri state3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Du, S, Deng, M.J, Xiao, J.Y. | | Deposit date: | 2024-05-09 | | Release date: | 2024-07-10 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Structural insights into the high-affinity IgE receptor Fc epsilon RI complex.
Nature, 633, 2024
|
|
1UWR
 
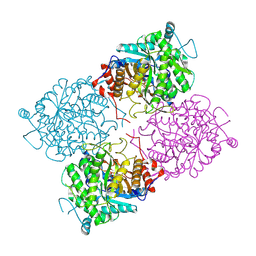 | | Structure of beta-glycosidase from Sulfolobus solfataricus in complex with 2-deoxy-2-fluoro-galactose | | Descriptor: | 2-deoxy-2-fluoro-alpha-D-galactopyranose, ACETATE ION, BETA-GALACTOSIDASE | | Authors: | Gloster, T.M, Roberts, S, Ducros, V.M.-A, Perugino, G, Rossi, M, Hoos, R, Moracci, M, Vasella, A, Davies, G.J. | | Deposit date: | 2004-02-11 | | Release date: | 2004-05-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Structural Studies of the Beta-Glycosidase from Sulfolobus Solfataricus in Complex with Covalently and Noncovalently Bound Inhibitors.
Biochemistry, 43, 2004
|
|
1V9L
 
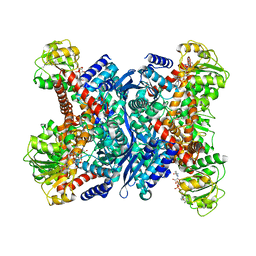 | | L-glutamate dehydrogenase from Pyrobaculum islandicum complexed with NAD | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, glutamate dehydrogenase | | Authors: | Bhuiya, M.W, Sakuraba, H, Ohshima, T, Imagawa, T, Katunuma, N, Tsuge, H. | | Deposit date: | 2004-01-26 | | Release date: | 2004-12-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The First Crystal Structure of Hyperthermostable NAD-dependent Glutamate Dehydrogenase from Pyrobaculum islandicum
J.Mol.Biol., 345, 2005
|
|
1VAT
 
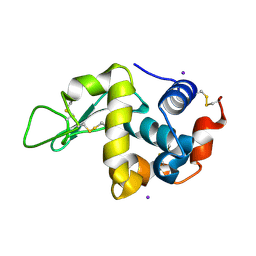 | | Iodine derivative of hen egg-white lysozyme | | Descriptor: | IODIDE ION, Lysozyme C | | Authors: | Takeda, K, Miyatake, H, Park, S.Y, Kawamoto, M, Kamiya, N, Miki, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-02-19 | | Release date: | 2005-03-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Multi-wavelength anomalous diffraction method for I and Xe atoms using ultra-high-energy X-rays from SPring-8
J.Appl.Crystallogr., 37, 2004
|
|
1B1Y
 
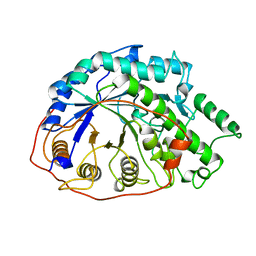 | | SEVENFOLD MUTANT OF BARLEY BETA-AMYLASE | | Descriptor: | PROTEIN (BETA-AMYLASE), alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose, beta-D-glucopyranose | | Authors: | Mikami, B, Yoon, H.J, Yoshigi, N. | | Deposit date: | 1998-11-25 | | Release date: | 1998-12-02 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of the sevenfold mutant of barley beta-amylase with increased thermostability at 2.5 A resolution.
J.Mol.Biol., 285, 1999
|
|
7BJL
 
 | | 14-3-3 sigma with RelA/p65 binding site pS45 and covalently bound TCF521-142 | | Descriptor: | 14-3-3 protein sigma, 3-[(3~{S})-3-methoxypiperidin-1-yl]-4-nitro-benzaldehyde, Transcription factor p65 | | Authors: | Wolter, M, Ottmann, C. | | Deposit date: | 2021-01-14 | | Release date: | 2021-06-09 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | An Exploration of Chemical Properties Required for Cooperative Stabilization of the 14-3-3 Interaction with NF-kappa B-Utilizing a Reversible Covalent Tethering Approach.
J.Med.Chem., 64, 2021
|
|
3EUH
 
 | | Crystal Structure of the MukE-MukF Complex | | Descriptor: | Chromosome partition protein mukF, GLYCINE, MukE | | Authors: | Suh, M.K, Ku, B, Ha, N.C, Woo, J.S, Oh, B.H. | | Deposit date: | 2008-10-10 | | Release date: | 2009-01-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural studies of a bacterial condensin complex reveal ATP-dependent disruption of intersubunit interactions.
Cell(Cambridge,Mass.), 136, 2009
|
|
