8F10
 
 | | Structure of the MDM2 P53 binding domain in complex with H102, an all-D Helicon Polypeptide | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, E3 ubiquitin-protein ligase Mdm2, ... | | Authors: | Li, K, Callahan, A.J, Travaline, T.L, Tokareva, O.S, Swiecicki, J.-M, Verdine, G.L, Pentelute, B.L, McGee, J.H. | | Deposit date: | 2022-11-04 | | Release date: | 2023-02-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Single-Shot Flow Synthesis of D-Proteins for Mirror-Image Phage Display
Chemrxiv, 2023
|
|
8F0Z
 
 | | Structure of the MDM2 P53 binding domain in complex with H101, an all-D Helicon Polypeptide | | Descriptor: | CHLORIDE ION, E3 ubiquitin-protein ligase Mdm2, H101, ... | | Authors: | Li, K, Callahan, A.J, Travaline, T.L, Tokareva, O.S, Swiecicki, J.-M, Verdine, G.L, Pentelute, B.L, McGee, J.H. | | Deposit date: | 2022-11-04 | | Release date: | 2023-02-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Single-Shot Flow Synthesis of D-Proteins for Mirror-Image Phage Display
Chemrxiv, 2023
|
|
5I8U
 
 | | Crystal Structure of the RV1700 (MT ADPRASE) E142Q mutant | | Descriptor: | ADP-ribose pyrophosphatase, DI(HYDROXYETHYL)ETHER, FORMIC ACID, ... | | Authors: | Thirawatananond, P, Kang, L.-W, Amzel, L.M, Gabelli, S.B. | | Deposit date: | 2016-02-19 | | Release date: | 2016-10-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Kinetic and mutational studies of the adenosine diphosphate ribose hydrolase from Mycobacterium tuberculosis.
J. Bioenerg. Biomembr., 48, 2016
|
|
7K6K
 
 | | Carbonic Anhydrase II complexed with 4-(2-(3-(3,5-dimethylphenyl)ureido)ethylsulfonamido)benzenesulfonamide | | Descriptor: | 4-{[(2-{[(3,5-dimethylphenyl)carbamoyl]amino}ethyl)sulfonyl]amino}benzene-1-sulfonamide, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | Authors: | Andring, J.T, Singh, S, McKenna, R. | | Deposit date: | 2020-09-20 | | Release date: | 2020-12-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.306 Å) | | Cite: | Handling drug-target selectivity: A study on ureido containing Carbonic Anhydrase inhibitors.
Eur.J.Med.Chem., 212, 2021
|
|
7K6T
 
 | | Carbonic Anhydrase IX mimic complexed with 4-(2-(3-phenylureido)ethylsulfonamido)benzenesulfonamide | | Descriptor: | 4-[({2-[(phenylcarbamoyl)amino]ethyl}sulfonyl)amino]benzene-1-sulfonamide, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Andring, J.T, Singh, S, McKenna, R. | | Deposit date: | 2020-09-21 | | Release date: | 2020-12-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.759 Å) | | Cite: | Handling drug-target selectivity: A study on ureido containing Carbonic Anhydrase inhibitors.
Eur.J.Med.Chem., 212, 2021
|
|
7BCQ
 
 | | ASCT2 in the presence of the inhibitor Lc-BPE (position "up") in the outward-open conformation. | | Descriptor: | 4-(4-phenylphenyl)carbonyloxypyrrolidine-2-carboxylic acid, Neutral amino acid transporter B(0) | | Authors: | Garibsingh, R.A, Ndaru, E, Garaeva, A.A, Shi, Y, Zielewicz, L, Bonomi, M, Slotboom, D.J, Paulino, C, Grewer, C, Schlessinger, A. | | Deposit date: | 2020-12-21 | | Release date: | 2021-09-22 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.43 Å) | | Cite: | Rational design of ASCT2 inhibitors using an integrated experimental-computational approach.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7BCT
 
 | | ASCT2 in the presence of the inhibitor ERA-21 in the outward-open conformation. | | Descriptor: | Neutral amino acid transporter B(0) | | Authors: | Garibsingh, R.A, Ndaru, E, Garaeva, A.A, Shi, Y, Zielewicz, L, Bonomi, M, Slotboom, D.J, Paulino, C, Grewer, C, Schlessinger, A. | | Deposit date: | 2020-12-21 | | Release date: | 2021-09-22 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Rational design of ASCT2 inhibitors using an integrated experimental-computational approach.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7BCS
 
 | | ASCT2 in the presence of the inhibitor Lc-BPE (position "down") in the outward-open conformation. | | Descriptor: | (2~{S},4~{S})-4-(4-phenylphenyl)carbonyloxypyrrolidine-2-carboxylic acid, Neutral amino acid transporter B(0) | | Authors: | Garibsingh, R.A, Ndaru, E, Garaeva, A.A, Shi, Y, Zielewicz, L, Bonomi, M, Slotboom, D.J, Paulino, C, Grewer, C, Schlessinger, A. | | Deposit date: | 2020-12-21 | | Release date: | 2021-09-22 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.43 Å) | | Cite: | Rational design of ASCT2 inhibitors using an integrated experimental-computational approach.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
5HBL
 
 | | Native rhodanese domain of YgaP prepared with 1mM DDT is S-nitrosylated | | Descriptor: | Inner membrane protein YgaP | | Authors: | Eichmann, C, Tzitzilonis, C, Nakamura, T, Kwiatkowski, W, Maslennikov, I, Choe, S, Lipton, S.A, Riek, R. | | Deposit date: | 2015-12-31 | | Release date: | 2016-08-10 | | Last modified: | 2021-09-08 | | Method: | X-RAY DIFFRACTION (1.617 Å) | | Cite: | S-Nitrosylation Induces Structural and Dynamical Changes in a Rhodanese Family Protein.
J.Mol.Biol., 428, 2016
|
|
7K6J
 
 | | Carbonic Anhydrase II complexed with 4-(2-(3-(4-methylphenyl)ureido)ethylsulfonamido)benzenesulfonamide | | Descriptor: | 4-{[(2-{[(4-methylphenyl)carbamoyl]amino}ethyl)sulfonyl]amino}benzene-1-sulfonamide, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | Authors: | Andring, J.T, Singh, S, McKenna, R. | | Deposit date: | 2020-09-20 | | Release date: | 2020-12-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.755 Å) | | Cite: | Handling drug-target selectivity: A study on ureido containing Carbonic Anhydrase inhibitors.
Eur.J.Med.Chem., 212, 2021
|
|
4O02
 
 | | AlphaVBeta3 integrin in complex with monoclonal antibody FAB fragment. | | Descriptor: | 17E6 heavy chain, 17E6 light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Mahalingam, B, van Agthoven, J, Xiong, J, Arnaout, M.A. | | Deposit date: | 2013-12-13 | | Release date: | 2014-04-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.605 Å) | | Cite: | Atomic basis for the species-specific inhibition of alpha V integrins by monoclonal antibody 17E6 is revealed by the crystal structure of alpha V beta 3 ectodomain-17E6 Fab complex.
J.Biol.Chem., 289, 2014
|
|
1BVI
 
 | | RIBONUCLEASE T1 (WILDTYPE) COMPLEXED WITH 2'GMP | | Descriptor: | CALCIUM ION, GUANOSINE-2'-MONOPHOSPHATE, PROTEIN (RIBONUCLEASE T1) | | Authors: | Langhorst, U, Loris, R, Denisov, V.P, Doumen, J, Roose, P, Maes, D, Halle, B, Steyaert, J. | | Deposit date: | 1998-09-15 | | Release date: | 1998-09-23 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Dissection of the structural and functional role of a conserved hydration site in RNase T1.
Protein Sci., 8, 1999
|
|
3K71
 
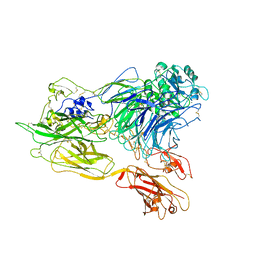 | | Structure of integrin alphaX beta2 ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Xie, C, Zhu, J, Chen, X, Mi, L, Nishida, N, Springer, T.A. | | Deposit date: | 2009-10-11 | | Release date: | 2010-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.95 Å) | | Cite: | Structure of an integrin with an alphaI domain, complement receptor type 4.
Embo J., 29, 2010
|
|
7K6U
 
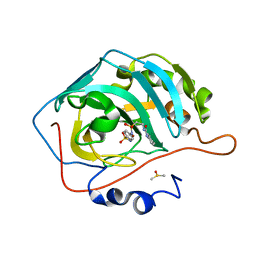 | | Carbonic Anhydrase IX mimic complexed with 4-(2-(3-(4-methylphenyl)ureido)ethylsulfonamido)benzenesulfonamide | | Descriptor: | 4-{[(2-{[(4-methylphenyl)carbamoyl]amino}ethyl)sulfonyl]amino}benzene-1-sulfonamide, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | Authors: | Andring, J.T, Singh, S, McKenna, R. | | Deposit date: | 2020-09-21 | | Release date: | 2020-12-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.605 Å) | | Cite: | Handling drug-target selectivity: A study on ureido containing Carbonic Anhydrase inhibitors.
Eur.J.Med.Chem., 212, 2021
|
|
7K6L
 
 | |
3SSI
 
 | |
5JQ5
 
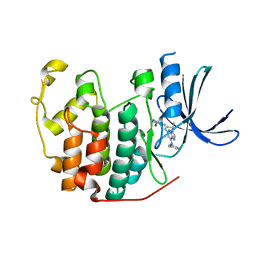 | | Crystal structure of CDK2 in complex with inhibitor ICEC0942 | | Descriptor: | (3R,4R)-4-[[[7-[(phenylmethyl)amino]-3-propan-2-yl-pyrazolo[1,5-a]pyrimidin-5-yl]amino]methyl]piperidin-3-ol, ACETATE ION, Cyclin-dependent kinase 2 | | Authors: | Hazel, P, Freemont, P.S. | | Deposit date: | 2016-05-04 | | Release date: | 2017-02-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Inhibitor Selectivity for Cyclin-Dependent Kinase 7: A Structural, Thermodynamic, and Modelling Study.
ChemMedChem, 12, 2017
|
|
5JQ8
 
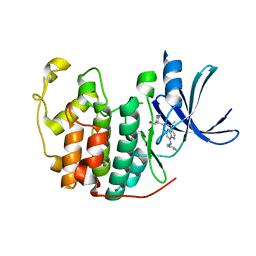 | | Crystal structure of CDK2 in complex with inhibitor ICEC0943 | | Descriptor: | (3S,4S)-4-[[[7-[(phenylmethyl)amino]-3-propan-2-yl-pyrazolo[1,5-a]pyrimidin-5-yl]amino]methyl]piperidin-3-ol, Cyclin-dependent kinase 2 | | Authors: | Hazel, P, Freemont, P.S. | | Deposit date: | 2016-05-04 | | Release date: | 2017-02-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Inhibitor Selectivity for Cyclin-Dependent Kinase 7: A Structural, Thermodynamic, and Modelling Study.
ChemMedChem, 12, 2017
|
|
1V6W
 
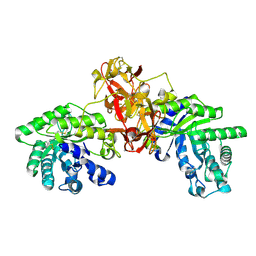 | | Crystal Structure Of Xylanase From Streptomyces Olivaceoviridis E-86 Complexed With 2(2)-4-O-methyl-alpha-D-glucuronosyl-xylobiose | | Descriptor: | 4-O-methyl-alpha-D-glucopyranuronic acid-(1-2)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose, ENDO-1,4-BETA-D-XYLANASE, beta-D-xylopyranose, ... | | Authors: | Fujimoto, Z, Kaneko, S, Kuno, A, Kobayashi, H, Kusakabe, I, Mizuno, H. | | Deposit date: | 2003-12-04 | | Release date: | 2004-04-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of decorated xylooligosaccharides bound to a family 10 xylanase from Streptomyces olivaceoviridis E-86
J.Biol.Chem., 279, 2004
|
|
8PP7
 
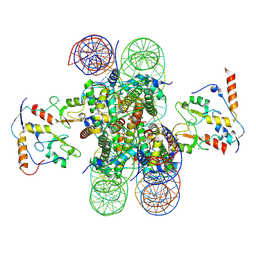 | | human RYBP-PRC1 bound to mononucleosome | | Descriptor: | DNA (215-mer), E3 ubiquitin-protein ligase RING2, Histone H2A, ... | | Authors: | Ciapponi, M, Benda, C, Mueller, J. | | Deposit date: | 2023-07-06 | | Release date: | 2024-03-27 | | Last modified: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | Structural basis of the histone ubiquitination read-write mechanism of RYBP-PRC1.
Nat.Struct.Mol.Biol., 31, 2024
|
|
3GS0
 
 | | Human transthyretin (TTR) complexed with (S)-3-(9H-fluoren-9-ylideneaminooxy)-2-methylpropanoic acid (inhibitor 16) | | Descriptor: | (2S)-3-[(9H-fluoren-9-ylideneamino)oxy]-2-methylpropanoic acid, Transthyretin | | Authors: | Mohamedmohaideen, N.N, Palaninathan, S.K, Orlandini, E, Sacchettini, J.C. | | Deposit date: | 2009-03-26 | | Release date: | 2009-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Novel transthyretin amyloid fibril formation inhibitors: synthesis, biological evaluation, and X-ray structural analysis.
Plos One, 4, 2009
|
|
7XWE
 
 | | RRGSGG-AtPRT6 UBR box | | Descriptor: | E3 ubiquitin-protein ligase PRT6, MAGNESIUM ION, ZINC ION | | Authors: | Kim, L, Song, H.K. | | Deposit date: | 2022-05-26 | | Release date: | 2023-05-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.598 Å) | | Cite: | Structural analyses of plant PRT6-UBR box for Cys-Arg/N-degron pathway and insights into the plant submergence resistance
To Be Published
|
|
7XWG
 
 | | RSGSGG-AtPRT6 UBR box | | Descriptor: | E3 ubiquitin-protein ligase PRT6, ZINC ION | | Authors: | Kim, L, Song, H.K. | | Deposit date: | 2022-05-26 | | Release date: | 2023-05-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.832 Å) | | Cite: | Structural analyses of plant PRT6-UBR box for Cys-Arg/N-degron pathway and insights into the plant submergence resistance
To Be Published
|
|
7XWF
 
 | | RLGSGG-AtPRT6 UBR box (highest resolution) | | Descriptor: | E3 ubiquitin-protein ligase PRT6, ZINC ION | | Authors: | Kim, L, Song, H.K. | | Deposit date: | 2022-05-26 | | Release date: | 2023-05-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.451 Å) | | Cite: | Structural analyses of plant PRT6-UBR box for Cys-Arg/N-degron pathway and insights into the plant submergence resistance
To Be Published
|
|
5OC8
 
 | | HDM2 (17-111, WILD TYPE) COMPLEXED WITH NVP-HDM201 AT 1.56A | | Descriptor: | (4~{S})-5-(5-chloranyl-1-methyl-2-oxidanylidene-pyridin-3-yl)-4-(4-chlorophenyl)-2-(2,4-dimethoxypyrimidin-5-yl)-3-propan-2-yl-4~{H}-pyrrolo[3,4-d]imidazol-6-one, CHLORIDE ION, E3 ubiquitin-protein ligase Mdm2 | | Authors: | Kallen, J. | | Deposit date: | 2017-06-29 | | Release date: | 2018-08-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Dose and Schedule Determine Distinct Molecular Mechanisms Underlying the Efficacy of the p53-MDM2 Inhibitor HDM201.
Cancer Res., 78, 2018
|
|
