5IBT
 
 | | UCA Fab (unbound) from 6515 Lineage | | Descriptor: | GLYCEROL, UCA Heavy Chain, UCA Light Chain | | Authors: | Raymond, D.D, Harrison, S.C. | | Deposit date: | 2016-02-22 | | Release date: | 2016-11-09 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Influenza immunization elicits antibodies specific for an egg-adapted vaccine strain.
Nat. Med., 22, 2016
|
|
5I2D
 
 | | Crystal structure of T. thermophilus TTHB099 class II transcription activation complex: TAP-RPo | | Descriptor: | DNA (72-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Feng, Y, Zhang, Y, Ebright, R.H. | | Deposit date: | 2016-02-08 | | Release date: | 2016-06-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (4.405 Å) | | Cite: | Structural basis of transcription activation.
Science, 352, 2016
|
|
3G2D
 
 | | Complex of Mth0212 and a 4 bp dsDNA with 3'-overhang | | Descriptor: | 5'-D(*CP*CP*TP*GP*UP*GP*CP*GP*AP*T)-3', 5'-D(*CP*GP*CP*G*CP*AP*GP*GP*C)-3', Exodeoxyribonuclease, ... | | Authors: | Lakomek, K, Dickmanns, A, Ficner, R. | | Deposit date: | 2009-01-31 | | Release date: | 2010-03-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure Analysis of DNA Uridine Endonuclease Mth212 Bound to DNA
J.Mol.Biol., 399, 2010
|
|
5IDP
 
 | | CDK8-CYCC IN COMPLEX WITH (3-Amino-1H-indazol-5-yl)-[(S)-2-(4-fluoro-phenyl)-piperidin-1-yl]-methanone | | Descriptor: | (3-amino-1H-indazol-5-yl)[(2S)-2-(4-fluorophenyl)piperidin-1-yl]methanone, Cyclin-C, Cyclin-dependent kinase 8, ... | | Authors: | Musil, D, Blagg, J, Mallinger, A, Czodrowski, P, Schiemann, K. | | Deposit date: | 2016-02-24 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure-Based Optimization of Potent, Selective, and Orally Bioavailable CDK8 Inhibitors Discovered by High-Throughput Screening.
J. Med. Chem., 59, 2016
|
|
5I6L
 
 | | Crystal Structure of Copper Nitrite Reductase at 100K after 2.76 MGy | | Descriptor: | ACETATE ION, COPPER (II) ION, Copper-containing nitrite reductase, ... | | Authors: | Horrell, S, Hough, M.A, Strange, R.W. | | Deposit date: | 2016-02-16 | | Release date: | 2016-07-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Serial crystallography captures enzyme catalysis in copper nitrite reductase at atomic resolution from one crystal.
Iucrj, 3, 2016
|
|
6EWT
 
 | | Solution Structure of Rhabdopeptide NRPS Docking Domain Kj12B-NDD | | Descriptor: | NRPS Kj12B-NDD | | Authors: | Hacker, C, Cai, X, Kegler, C, Zhao, L, Weickhmann, A.K, Bode, H.B, Woehnert, J. | | Deposit date: | 2017-11-06 | | Release date: | 2018-10-31 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structure-based redesign of docking domain interactions modulates the product spectrum of a rhabdopeptide-synthesizing NRPS.
Nat Commun, 9, 2018
|
|
6BM4
 
 | | Pol II elongation complex with an abasic lesion at i-1 position,soaking UMPNPP | | Descriptor: | 5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]uridine, DNA (5'-D(P*CP*AP*(3DR)P*CP*TP*CP*TP*TP*GP*AP*TP*G)-3'), DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Wang, W, Wang, D. | | Deposit date: | 2017-11-13 | | Release date: | 2018-02-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.951 Å) | | Cite: | Structural basis of transcriptional stalling and bypass of abasic DNA lesion by RNA polymerase II.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6BNO
 
 | | Structure of bare actin filament | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Gurel, P.S, Alushin, G.A. | | Deposit date: | 2017-11-17 | | Release date: | 2018-01-10 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (5.5 Å) | | Cite: | Cryo-EM structures reveal specialization at the myosin VI-actin interface and a mechanism of force sensitivity.
Elife, 6, 2017
|
|
1HB9
 
 | | quasi-atomic resolution model of bacteriophage PRD1 wild type virion, obtained by combined cryo-EM and X-ray crystallography. | | Descriptor: | BACTERIOPHAGE PRD1 | | Authors: | San Martin, C, Burnett, R.M, De Haas, F, Heinkel, R, Rutten, T, Fuller, S.D, Butcher, S.J, Bamford, D.H. | | Deposit date: | 2001-04-13 | | Release date: | 2001-12-05 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (25 Å) | | Cite: | Combined Em/X-Ray Imaging Yields a Quasi-Atomic Model of the Adenovirus-Related Bacteriophage Prd1 and Shows Key Capsid and Membrane Interactions
Structure, 9, 2001
|
|
5IYB
 
 | | Human core-PIC in the open state | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB10, DNA-directed RNA polymerase II subunit RPB11-a, ... | | Authors: | He, Y, Yan, C, Fang, J, Inouye, C, Tjian, R, Ivanov, I, Nogales, E. | | Deposit date: | 2016-03-24 | | Release date: | 2016-05-18 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Near-atomic resolution visualization of human transcription promoter opening.
Nature, 533, 2016
|
|
5JBC
 
 | |
5IYD
 
 | | Human core-PIC in the initial transcribing state (no IIS) | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB10, DNA-directed RNA polymerase II subunit RPB11-a, ... | | Authors: | He, Y, Yan, C, Fang, J, Inouye, C, Tjian, R, Ivanov, I, Nogales, E. | | Deposit date: | 2016-03-24 | | Release date: | 2016-05-18 | | Last modified: | 2019-11-20 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Near-atomic resolution visualization of human transcription promoter opening.
Nature, 533, 2016
|
|
6C98
 
 | | Crystal structure of FcRn bound to UCB-84 | | Descriptor: | 1-[7-(3-fluorophenyl)-5-methyl[1,2,4]triazolo[1,5-a]pyrimidin-6-yl]ethan-1-one, Beta-2-microglobulin, CYSTEINE, ... | | Authors: | Fox III, D, Lukacs, C.M. | | Deposit date: | 2018-01-25 | | Release date: | 2018-05-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Insight into small molecule binding to the neonatal Fc receptor by X-ray crystallography and 100 kHz magic-angle-spinning NMR.
PLoS Biol., 16, 2018
|
|
5J0D
 
 | | Crystal structure of the bromodomain of human CREBBP in complex with a benzoxazepine compound | | Descriptor: | 1,2-ETHANEDIOL, 7-(3,5-dimethoxyphenyl)-N-[(3S)-1-methylpiperidin-3-yl]-4-propanoyl-2,3,4,5-tetrahydro-1,4-benzoxazepine-9-carboxamide, CREB-binding protein | | Authors: | Tallant, C, Popp, T.A, Fedorov, O, Siejka, P, Picaud, S, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bracher, F, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-03-28 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Development of Selective CBP/P300 Benzoxazepine Bromodomain Inhibitors.
J.Med.Chem., 59, 2016
|
|
2NZ0
 
 | | Crystal structure of potassium channel Kv4.3 in complex with its regulatory subunit KChIP1 | | Descriptor: | CALCIUM ION, Kv channel-interacting protein 1, Potassium voltage-gated channel subfamily D member 3, ... | | Authors: | Wang, H, Yan, Y, Shen, Y, Chen, L, Wang, K. | | Deposit date: | 2006-11-22 | | Release date: | 2006-12-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for modulation of Kv4 K(+) channels by auxiliary KChIP subunits.
Nat.Neurosci., 10, 2007
|
|
6B8N
 
 | |
3FWQ
 
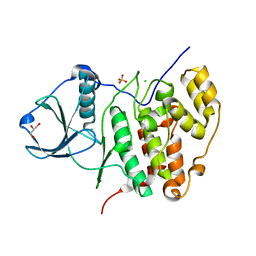 | | Inactive conformation of human protein kinase CK2 catalytic subunit | | Descriptor: | CHLORIDE ION, Casein kinase II subunit alpha, GLYCEROL, ... | | Authors: | Niefind, K, Raaf, J, Issinger, O.G. | | Deposit date: | 2009-01-19 | | Release date: | 2009-02-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | First inactive conformation of CK2 alpha, the catalytic subunit of protein kinase CK2
J.Mol.Biol., 386, 2009
|
|
5JBA
 
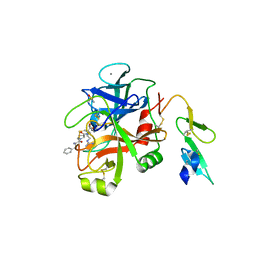 | |
5J1L
 
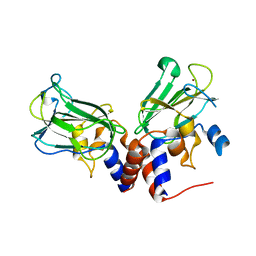 | | Crystal structure of Csd1-Csd2 dimer I | | Descriptor: | ToxR-activated gene (TagE), ZINC ION | | Authors: | An, D.R, Suh, S.W. | | Deposit date: | 2016-03-29 | | Release date: | 2016-10-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structural Basis of the Heterodimer Formation between Cell Shape-Determining Proteins Csd1 and Csd2 from Helicobacter pylori
Plos One, 11, 2016
|
|
5NEY
 
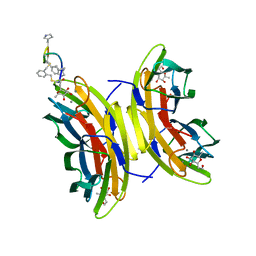 | | Discovery, crystal structures and atomic force microscopy study of thioether ligated D,L-cyclic antimicrobial peptides against multidrug resistant Pseudomonas aeruginosa | | Descriptor: | 3,7-anhydro-2,8-dideoxy-L-glycero-D-gluco-octonic acid, CALCIUM ION, CYS-TRD-TRP-LYD-LYS-LYD-LYS-LYD-TRP-TRD-CYS-ALA, ... | | Authors: | Reymond, J.-L, Darbre, T, Stocker, A, Hong, W, van Delden, C, Koehler, T, Luscher, A, Visini, R, Fu, Y, Di Bonaventura, I, He, R. | | Deposit date: | 2017-03-13 | | Release date: | 2017-09-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Design, crystal structure and atomic force microscopy study of thioether ligated d,l-cyclic antimicrobial peptides against multidrug resistant Pseudomonas aeruginosa.
Chem Sci, 8, 2017
|
|
6B8L
 
 | |
5JA5
 
 | | Crystal structure of the rice Topless related protein 2 (TPR2) N-terminal topless domain (1-209) L111A and L130A mutant in complex with rice D53 repressor EAR peptide motif | | Descriptor: | Protein TPR1, The rice D53 peptide (a.a. 794-808), ZINC ION | | Authors: | Ke, J, Ma, H, Gu, X, Brunzelle, J.S, Xu, H.E, Melcher, K. | | Deposit date: | 2016-04-12 | | Release date: | 2017-07-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A D53 repression motif induces oligomerization of TOPLESS corepressors and promotes assembly of a corepressor-nucleosome complex.
Sci Adv, 3, 2017
|
|
6B9M
 
 | |
2C8G
 
 | | Structure of the PN loop Q182A mutant C3bot1 Exoenzyme (Free state, crystal form I) | | Descriptor: | MONO-ADP-RIBOSYLTRANSFERASE C3, SULFATE ION | | Authors: | Stura, E.A, Menetrey, J, Flatau, G, Boquet, P, Menez, A. | | Deposit date: | 2005-12-03 | | Release date: | 2007-02-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Properties of Wild-Type and Two Artt Motif Mutants Clostridium Botulinum C3 Exoenzyme Isoform 1 in Different Substrate Complexed States and Crystal Forms.
To be Published
|
|
1F85
 
 | | SOLUTION STRUCTURE OF HCV IRES RNA DOMAIN IIIE | | Descriptor: | HCV-1B IRES RNA DOMAIN IIIE | | Authors: | Lukavsky, P.J, Otto, G.A, Lancaster, A.M, Sarnow, P, Puglisi, J.D. | | Deposit date: | 2000-06-28 | | Release date: | 2000-11-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structures of two RNA domains essential for hepatitis C virus internal ribosome entry site function.
Nat.Struct.Biol., 7, 2000
|
|
