7NIT
 
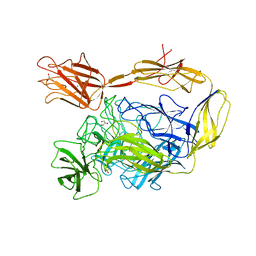 | | X-ray structure of a multidomain BbgIII from Bifidobacterium bifidum | | Descriptor: | Beta-galactosidase, CALCIUM ION, GLYCEROL, ... | | Authors: | Moroz, O.V, Blagova, E, Lebedev, A.A, Sanchez Rodriguez, F, Rigden, D.J, Tams, J.W, Wilting, R, Vester, J.K, Longhin, E, Krogh, K.B.R, Pache, R.A, Davies, G.J, Wilson, K.S. | | Deposit date: | 2021-02-14 | | Release date: | 2021-12-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Multitasking in the gut: the X-ray structure of the multidomain BbgIII from Bifidobacterium bifidum offers possible explanations for its alternative functions.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
6HDW
 
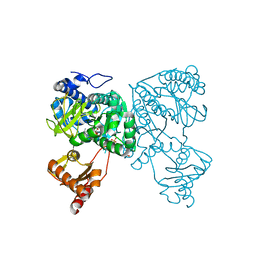 | | Crystal structure of 2-Hydroxyisobutyryl-CoA Ligase (HCL) in the postadenylation state in complex with 2-HIB-AMP | | Descriptor: | 2-hydroxyisobutyryl-CoA synthetase, SULFATE ION, [[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] 2-methyl-2-oxidanyl-propanoate | | Authors: | Zahn, M, Rohwerder, T, Strater, N. | | Deposit date: | 2018-08-20 | | Release date: | 2019-08-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of 2-Hydroxyisobutyric Acid-CoA Ligase Reveal Determinants of Substrate Specificity and Describe a Multi-Conformational Catalytic Cycle.
J.Mol.Biol., 431, 2019
|
|
7C22
 
 | |
6HJA
 
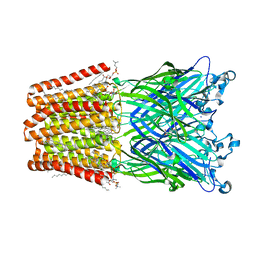 | | Xray structure of GLIC in complex with glutarate | | Descriptor: | CHLORIDE ION, DIUNDECYL PHOSPHATIDYL CHOLINE, DODECANE, ... | | Authors: | Fourati, Z, Delarue, M. | | Deposit date: | 2018-09-03 | | Release date: | 2019-09-18 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural evidence for the binding of monocarboxylates and dicarboxylates at pharmacologically relevant extracellular sites of a pentameric ligand-gated ion channel.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
8TXA
 
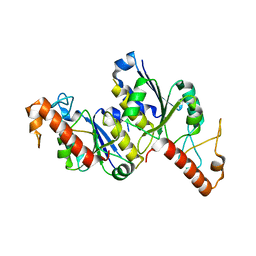 | | Apo Structure of (N1G37) tRNA Methyltransferase from Mycobacterium marinum | | Descriptor: | tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Balsamo, A, Bruno, C, Edele, C, Fabian, T, Jannotta, R, Lee, R, Schryver, D, Warsaw, J, Warsaw, L, Stojanoff, V, Battaile, K, Perez, A, Bolen, R. | | Deposit date: | 2023-08-23 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.591 Å) | | Cite: | Apo Structure of (N1G37) tRNA Methyltransferase from Mycobacterium marinum
To Be Published
|
|
7NHF
 
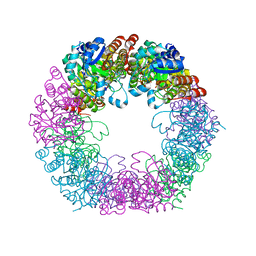 | | Crystal structure of Arabidopsis thaliana Pdx1K166R | | Descriptor: | PHOSPHATE ION, Pyridoxal 5'-phosphate synthase subunit PDX1.3 | | Authors: | Rodrigues, M.J, Zhang, Y, Bolton, R, Evans, G, Giri, N, Royant, A, Begley, T, Ealick, S.E, Tews, I. | | Deposit date: | 2021-02-10 | | Release date: | 2021-12-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Trapping and structural characterisation of a covalent intermediate in vitamin B 6 biosynthesis catalysed by the Pdx1 PLP synthase.
Rsc Chem Biol, 3, 2022
|
|
6BW3
 
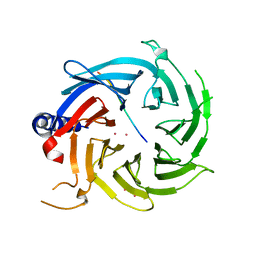 | | Crystal structure of RBBP4 in complex with PRDM3 N-terminal peptide | | Descriptor: | Histone-binding protein RBBP4, MDS1 and EVI1 complex locus protein MDS1, UNKNOWN ATOM OR ION | | Authors: | Ivanochko, D, Halabelian, L, Hutchinson, A, Seitova, A, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-12-14 | | Release date: | 2017-12-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Direct interaction between the PRDM3 and PRDM16 tumor suppressors and the NuRD chromatin remodeling complex.
Nucleic Acids Res., 47, 2019
|
|
6HEP
 
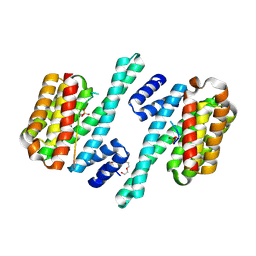 | |
6HEW
 
 | | Crystal Structure of Ephrin A2 (EphA2) Receptor Protein Kinase with the NVP-BHG712 derivative AT069 | | Descriptor: | 4-methyl-3-[(1-methyl-6-pyrimidin-5-yl-pyrazolo[3,4-d]pyrimidin-4-yl)amino]-~{N}-[3-(trifluoromethyl)phenyl]benzamide, Ephrin type-A receptor 2 | | Authors: | Kudlinzki, D, Troester, A, Witt, K, Linhard, V.L, Gande, S.L, Saxena, K, Schwalbe, H. | | Deposit date: | 2018-08-20 | | Release date: | 2019-08-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.268 Å) | | Cite: | Effects of NVP-BHG712 chemical modifications on EPHA2 binding and affinity
To Be Published
|
|
7MWI
 
 | | Crystal structure of human BAZ2A | | Descriptor: | Bromodomain adjacent to zinc finger domain protein 2A, UNKNOWN ATOM OR ION | | Authors: | Liu, K, Dong, A, Li, Y, Loppnau, P, Edwards, A.M, Arrowsmith, C.H, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-05-17 | | Release date: | 2021-12-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of the TAM domain of BAZ2A in binding to DNA or RNA independent of methylation status.
J.Biol.Chem., 297, 2021
|
|
8TNV
 
 | | Hemocyanin Functional Unit CCHB-g of Concholepas concholepas | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Munoz, S, Vallejos-Baccelliere, G, Manubens, A, Salazar, M, Nascimento, A.F.Z, Ambrosio, A.L.B, Becker, M.I, Guixe, V, Castro-Fernandez, V. | | Deposit date: | 2023-08-02 | | Release date: | 2024-04-10 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural insights into a functional unit from an immunogenic mollusk hemocyanin.
Structure, 32, 2024
|
|
7BVK
 
 | |
8TNT
 
 | | Crystal structure of Epstein-Barr virus gH/gL/gp42 in complex with antibodies F-2-1 and 769C2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 769C2 heavy chain, 769C2 light chain, ... | | Authors: | Bu, W, Kumar, A, Board, N, Kim, J, Dowdell, K, Zhang, S, Lei, Y, Hostal, A, Krogmann, T, Wang, Y, Pittaluga, S, Marcotrigiano, J, Cohen, J.I. | | Deposit date: | 2023-08-02 | | Release date: | 2024-03-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Epstein-Barr virus gp42 antibodies reveal sites of vulnerability for receptor binding and fusion to B cells.
Immunity, 57, 2024
|
|
7RTD
 
 | |
7S0L
 
 | |
8TQV
 
 | | Crystal Structure of Mtb Pks13 Thioesterase domain in complex with inhibitor X20403 | | Descriptor: | 4-(2-{(4M)-4-[(6M)-6-(2,5-dimethoxyphenyl)pyridin-3-yl]-1H-1,2,3-triazol-1-yl}ethyl)-N-{[1-(methoxymethyl)cyclopropyl]methyl}-N-methylbenzamide, Polyketide synthase Pks13, SULFATE ION | | Authors: | Krieger, I.V, Sacchettini, J.C. | | Deposit date: | 2023-08-08 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Inhibitors of the Thioesterase Activity of Mycobacterium tuberculosis Pks13 Discovered Using DNA-Encoded Chemical Library Screening.
Acs Infect Dis., 10, 2024
|
|
7S7V
 
 | | Crystal structure of iNicSnFR3a Fluorescent Nicotine Sensor | | Descriptor: | iNicSnFR 3.0 Fluorescent Nicotine Sensor | | Authors: | Fan, C, Shivange, A.V, Looger, L.L, Lester, H.A, Rees, D.C. | | Deposit date: | 2021-09-17 | | Release date: | 2021-10-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Correction: Fluorescence activation mechanism and imaging of drug permeation with new sensors for smoking-cessation ligands.
Elife, 11, 2022
|
|
7NNE
 
 | |
7BF7
 
 | |
7RTS
 
 | |
7NMA
 
 | |
7BGA
 
 | |
7RX1
 
 | |
6HG7
 
 | | Crystal structure of a collagen II fragment containing the binding site of PEDF and COMP, (POG)4-LKG HRG FTG LQG-POG(4) | | Descriptor: | Collagen alpha-1(II) chain, SULFATE ION | | Authors: | Gebauer, J.M, Koehler, A, Dietmar, H, Gompert, M, Neundorf, I, Zaucke, F, Koch, M, Baumann, U. | | Deposit date: | 2018-08-22 | | Release date: | 2018-12-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | COMP and TSP-4 interact specifically with the novel GXKGHR motif only found in fibrillar collagens.
Sci Rep, 8, 2018
|
|
7S7Z
 
 | | Crystal structure of iAChSnFR Acetylcholine Sensor precursor binding protein with choline bound | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, CHOLINE ION, iAchSnFR Fluorescent Acetylcholine Sensor precursor binding protein | | Authors: | Fan, C, Borden, P.M, Looger, L.L, Lester, H.A, Rees, D.C. | | Deposit date: | 2021-09-17 | | Release date: | 2021-10-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure, Function, and Application of Bacterial ABC Transporters
Ph.D.Thesis,California Institute of Technology, 2020
|
|
