1TKX
 
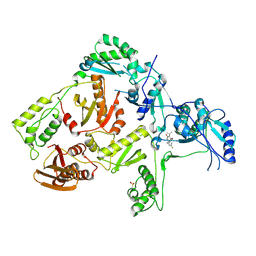 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH GW490745 | | Descriptor: | 4-[(CYCLOPROPYLETHYNYL)OXY]-6-FLUORO-3-ISOPROPYLQUINOLIN-2(1H)-ONE, Pol polyprotein, Reverse transcriptase, ... | | Authors: | Ren, J, Hopkins, A.L, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2004-06-09 | | Release date: | 2004-12-07 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Design of non-nucleoside inhibitors of HIV-1 reverse transcriptase with improved drug resistance properties. 2.
J.Med.Chem., 47, 2004
|
|
1JTM
 
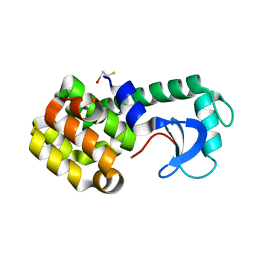 | |
1E9G
 
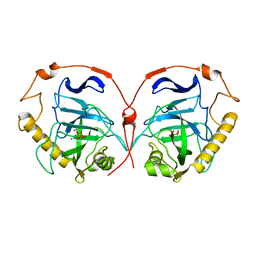 | | STRUCTURE OF INORGANIC PYROPHOSPHATASE | | Descriptor: | INORGANIC PYROPHOSPHATASE, MANGANESE (II) ION, PHOSPHATE ION | | Authors: | Heikinheimo, P, Tuominen, V, Ahonen, A.-K, Teplyakov, A, Cooperman, B.S, Baykov, A.A, Lahti, R, Goldman, A. | | Deposit date: | 2000-10-12 | | Release date: | 2001-03-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Toward a Quantum-Mechanical Description of Metal-Assisted Phosphoryl Transfer in Pyrophosphatase
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
2J8C
 
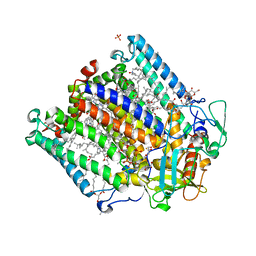 | | X-ray high resolution structure of the photosynthetic reaction center from Rb. sphaeroides at pH 8 in the neutral state | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, ... | | Authors: | Koepke, J, Diehm, R, Fritzsch, G. | | Deposit date: | 2006-10-24 | | Release date: | 2007-07-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Ph Modulates the Quinone Position in the Photosynthetic Reaction Center from Rhodobacter Sphaeroides in the Neutral and Charge Separated States.
J.Mol.Biol., 371, 2007
|
|
1JRU
 
 | | NMR STRUCTURE OF THE UBX DOMAIN FROM P47 (ENERGY MINIMISED AVERAGE) | | Descriptor: | p47 protein | | Authors: | Yuan, X.M, Shaw, A, Zhang, X.D, Kondo, H, Lally, J, Freemont, P.S, Matthews, S.J. | | Deposit date: | 2001-08-15 | | Release date: | 2001-08-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure and interaction surface of the C-terminal domain from p47: a major p97-cofactor involved in SNARE disassembly.
J.Mol.Biol., 311, 2001
|
|
1E6A
 
 | | Fluoride-inhibited substrate complex of Saccharomyces cerevisiae inorganic pyrophosphatase | | Descriptor: | FLUORIDE ION, INORGANIC PYROPHOSPHATASE, MANGANESE (II) ION, ... | | Authors: | Heikinheimo, P, Tuominen, V, Ahonen, A.-K, Teplyakov, A, Cooperman, B.S, Baykov, A.A, Lahti, R, Goldman, A. | | Deposit date: | 2000-08-09 | | Release date: | 2001-03-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Toward a quantum-mechanical description of metal-assisted phosphoryl transfer in pyrophosphatase.
Proc. Natl. Acad. Sci. U.S.A., 98, 2001
|
|
1A80
 
 | | Native 2,5-DIKETO-D-GLUCONIC acid reductase a from CORYNBACTERIUM SP. complexed with nadph | | Descriptor: | 2,5-DIKETO-D-GLUCONIC ACID REDUCTASE A, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Khurana, S, Powers, D.B, Anderson, S, Blaber, M. | | Deposit date: | 1998-03-31 | | Release date: | 1999-03-30 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of 2,5-diketo-D-gluconic acid reductase A complexed with NADPH at 2.1-A resolution.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1EAP
 
 | | CRYSTAL STRUCTURE OF A CATALYTIC ANTIBODY WITH A SERINE PROTEASE ACTIVE SITE | | Descriptor: | IGG2B-KAPPA 17E8 FAB (HEAVY CHAIN), IGG2B-KAPPA 17E8 FAB (LIGHT CHAIN), PHENYL[1-(N-SUCCINYLAMINO)PENTYL]PHOSPHONATE | | Authors: | Zhou, G.W, Guo, J, Huang, W, Scanlan, T.S, Fletterick, R.J. | | Deposit date: | 1994-08-10 | | Release date: | 1994-12-20 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of a catalytic antibody with a serine protease active site.
Science, 265, 1994
|
|
1UA7
 
 | | Crystal Structure Analysis of Alpha-Amylase from Bacillus Subtilis complexed with Acarbose | | Descriptor: | 4,6-dideoxy-alpha-D-xylo-hexopyranose-(1-4)-alpha-D-glucopyranose, 6-AMINO-4-HYDROXYMETHYL-CYCLOHEX-4-ENE-1,2,3-TRIOL, Alpha-amylase, ... | | Authors: | Kagawa, M, Fujimoto, Z, Momma, M, Takase, K, Mizuno, H. | | Deposit date: | 2003-03-03 | | Release date: | 2004-05-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Crystal structure of Bacillus subtilis alpha-amylase in complex with acarbose
J.BACTERIOL., 185, 2003
|
|
1JXC
 
 | | Minimized NMR structure of ATT, an Arabidopsis trypsin/chymotrypsin inhibitor | | Descriptor: | Putative trypsin inhibitor ATTI-2 | | Authors: | Zhao, Q, Chae, Y.K, Markley, J.L, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2001-09-06 | | Release date: | 2003-01-07 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | NMR Solution Structure of ATTp, an Arabidopsis thaliana Trypsin Inhibitor
Biochemistry, 41, 2002
|
|
1UDW
 
 | | Crystal structure of human uridine-cytidine kinase 2 complexed with a feedback-inhibitor, CTP | | Descriptor: | CYTIDINE-5'-TRIPHOSPHATE, Uridine-cytidine kinase 2 | | Authors: | Suzuki, N.N, Koizumi, K, Fukushima, M, Matsuda, A, Inagaki, F. | | Deposit date: | 2003-05-07 | | Release date: | 2004-05-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for the specificity, catalysis, and regulation of human uridine-cytidine kinase
STRUCTURE, 12, 2004
|
|
1F0P
 
 | | MYCOBACTERIUM TUBERCULOSIS ANTIGEN 85B WITH TREHALOSE | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ANTIGEN 85-B, ... | | Authors: | Anderson, D.H, Harth, G, Horwitz, M.A, Eisenberg, D, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2000-05-16 | | Release date: | 2001-01-24 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | An interfacial mechanism and a class of inhibitors inferred from two crystal structures of the Mycobacterium tuberculosis 30 kDa major secretory protein (Antigen 85B), a mycolyl transferase.
J.Mol.Biol., 307, 2001
|
|
1XLD
 
 | | MECHANISM FOR ALDOSE-KETOSE INTERCONVERSION BY D-XYLOSE ISOMERASE INVOLVING RING OPENING FOLLOWED BY A 1,2-HYDRIDE SHIFT | | Descriptor: | D-XYLOSE ISOMERASE, MANGANESE (II) ION, Xylitol | | Authors: | Collyer, C.A, Henrick, K, Blow, D.M. | | Deposit date: | 1991-10-09 | | Release date: | 1993-07-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanism for aldose-ketose interconversion by D-xylose isomerase involving ring opening followed by a 1,2-hydride shift.
J.Mol.Biol., 212, 1990
|
|
1A12
 
 | | REGULATOR OF CHROMOSOME CONDENSATION (RCC1) OF HUMAN | | Descriptor: | REGULATOR OF CHROMOSOME CONDENSATION 1 | | Authors: | Renault, L, Nassar, N, Vetter, I, Becker, J, Roth, M, Wittinghofer, A. | | Deposit date: | 1997-12-19 | | Release date: | 1999-01-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The 1.7 A crystal structure of the regulator of chromosome condensation (RCC1) reveals a seven-bladed propeller.
Nature, 392, 1998
|
|
1A6M
 
 | | OXY-MYOGLOBIN, ATOMIC RESOLUTION | | Descriptor: | MYOGLOBIN, OXYGEN MOLECULE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Vojtechovsky, J, Chu, K, Berendzen, J, Sweet, R.M, Schlichting, I. | | Deposit date: | 1998-02-26 | | Release date: | 1999-04-06 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Crystal structures of myoglobin-ligand complexes at near-atomic resolution.
Biophys.J., 77, 1999
|
|
1U3I
 
 | | Crystal structure of glutathione S-tranferase from Schistosoma mansoni | | Descriptor: | GLUTATHIONE, Glutathione S-transferase 28 kDa | | Authors: | Chomilier, J, Vaney, M.C, Labesse, G, Trottein, F, Capron, A, Mormon, J.-P. | | Deposit date: | 2004-07-22 | | Release date: | 2005-07-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal structure of Schistosoma mansoni glutathione S-transferase
To be Published
|
|
1TVQ
 
 | | Crystal Structure of Apo Chicken Liver Basic Fatty Acid Binding Protein (or Bile Acid Binding Protein) | | Descriptor: | Fatty acid-binding protein | | Authors: | Nichesola, D, Perduca, M, Capaldi, S, Carrizo, M.E, Righetti, P.G, Monaco, H.L. | | Deposit date: | 2004-06-30 | | Release date: | 2004-11-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of chicken liver basic Fatty Acid-binding protein complexed with cholic acid
Biochemistry, 43, 2004
|
|
1TW9
 
 | | Glutathione Transferase-2, apo form, from the nematode Heligmosomoides polygyrus | | Descriptor: | glutathione S-transferase 2 | | Authors: | Schuller, D.J, Liu, Q, Kriksunov, I.A, Campbell, A.M, Barrett, J, Brophy, P.M, Hao, Q. | | Deposit date: | 2004-06-30 | | Release date: | 2004-08-03 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Crystal structure of a new class of glutathione transferase from the model human hookworm nematode Heligmosomoides polygyrus.
Proteins, 61, 2005
|
|
1AG3
 
 | |
1XIG
 
 | |
1F0N
 
 | | MYCOBACTERIUM TUBERCULOSIS ANTIGEN 85B | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ANTIGEN 85B | | Authors: | Anderson, D.H, Harth, G, Horwitz, M.A, Eisenberg, D, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2000-05-16 | | Release date: | 2001-01-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | An interfacial mechanism and a class of inhibitors inferred from two crystal structures of the Mycobacterium tuberculosis 30 kDa major secretory protein (Antigen 85B), a mycolyl transferase.
J.Mol.Biol., 307, 2001
|
|
1F08
 
 | | CRYSTAL STRUCTURE OF THE DNA-BINDING DOMAIN OF THE REPLICATION INITIATION PROTEIN E1 FROM PAPILLOMAVIRUS | | Descriptor: | BROMIDE ION, REPLICATION PROTEIN E1 | | Authors: | Enemark, E.J, Chen, G, Vaughn, D.E, Stenlund, A, Joshua-Tor, L. | | Deposit date: | 2000-05-15 | | Release date: | 2001-05-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the DNA binding domain of the replication initiation protein E1 from papillomavirus.
Mol.Cell, 6, 2000
|
|
1XIH
 
 | |
1TO4
 
 | | Structure of the cytosolic Cu,Zn SOD from S. mansoni | | Descriptor: | COPPER (II) ION, Superoxide dismutase, ZINC ION | | Authors: | Cardoso, R.M.F, Silva, C.H.T.P, Ulian de Araujo, A.P, Tanaka, T, Tanaka, M, Garratt, R.C. | | Deposit date: | 2004-06-12 | | Release date: | 2004-08-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure of the cytosolic Cu,Zn superoxide dismutase from Schistosoma mansoni.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1F1H
 
 | |
