2Z69
 
 | |
3DEX
 
 | | Crystal structure of SAV_2001 protein from Streptomyces avermitilis, Northeast Structural Genomics Consortium Target SvR107. | | Descriptor: | SAV_2001 | | Authors: | Forouhar, F, Neely, H, Seetharaman, J, Janjua, H, Fang, Y, Xiao, R, Cunningham, K, Ma, L.-C, Owen, L.A, Chen, C.X, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-06-10 | | Release date: | 2008-08-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of SAV_2001 protein from Streptomyces avermitilis, Northeast Structural Genomics Consortium Target SvR107.
To be Published
|
|
7GTJ
 
 | |
4KHM
 
 | | HCV NS5B GT1A with GSK5852 | | Descriptor: | HCV Polymerase, SULFATE ION, [4-({[5-cyclopropyl-2-(4-fluorophenyl)-3-(methylcarbamoyl)-1-benzofuran-6-yl](methylsulfonyl)amino}methyl)-2-fluorophenyl]boronic acid | | Authors: | Williams, S.P, Kahler, K.M, Shotwell, J.B. | | Deposit date: | 2013-04-30 | | Release date: | 2013-05-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of a Potent Boronic Acid Derived Inhibitor of the HCV RNA-Dependent RNA Polymerase.
J.Med.Chem., 57, 2014
|
|
1M4L
 
 | | STRUCTURE OF NATIVE CARBOXYPEPTIDASE A AT 1.25 RESOLUTION | | Descriptor: | CARBOXYPEPTIDASE A, ZINC ION | | Authors: | Kilshtain-Vardi, A, Glick, M, Greenblatt, H.M, Goldblum, A, Shoham, G. | | Deposit date: | 2002-07-03 | | Release date: | 2003-01-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Refined structure of bovine carboxypeptidase A at 1.25 A resolution.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
7PP6
 
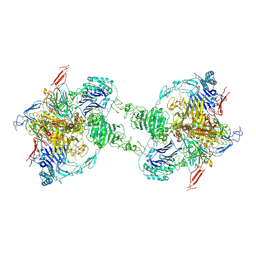 | | MUC2 Tubules of D1D2D3 domains | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Javitt, G, Fass, D. | | Deposit date: | 2021-09-13 | | Release date: | 2022-02-16 | | Last modified: | 2022-05-25 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Helical self-assembly of a mucin segment suggests an evolutionary origin for von Willebrand factor tubules.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7GTE
 
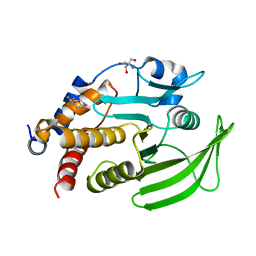 | | PanDDA Analysis group deposition -- Crystal structure of PTP1B in complex with XST00000646b | | Descriptor: | (5S)-5-(trifluoromethyl)-1,4-diazepane, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Mehlman, T, Ginn, H.M, Keedy, D.A. | | Deposit date: | 2024-01-03 | | Release date: | 2024-01-24 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | An expanded view of ligandability in the allosteric enzyme PTP1B from computational reanalysis of large-scale crystallographic data.
Biorxiv, 2024
|
|
7GTM
 
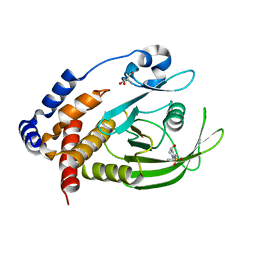 | | PanDDA Analysis group deposition -- Crystal structure of PTP1B in complex with FMOOA000543a | | Descriptor: | (4S)-4-hydroxy-2-(propan-2-yl)-3,4-dihydro-1lambda~6~,2-benzothiazine-1,1(2H)-dione, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Mehlman, T, Ginn, H.M, Keedy, D.A. | | Deposit date: | 2024-01-03 | | Release date: | 2024-01-24 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | An expanded view of ligandability in the allosteric enzyme PTP1B from computational reanalysis of large-scale crystallographic data.
Biorxiv, 2024
|
|
1M4Z
 
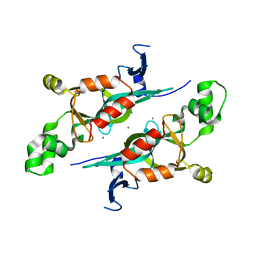 | | Crystal structure of the N-terminal BAH domain of Orc1p | | Descriptor: | MANGANESE (II) ION, ORIGIN RECOGNITION COMPLEX SUBUNIT 1 | | Authors: | Zhang, Z, Hayashi, M.K, Merkel, O, Stillman, B, Xu, R.-M. | | Deposit date: | 2002-07-05 | | Release date: | 2002-09-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and function of the BAH-containing domain of Orc1p in epigenetic silencing.
EMBO J., 21, 2002
|
|
7GTV
 
 | |
4KIV
 
 | |
7GS8
 
 | | PanDDA Analysis group deposition -- Crystal structure of PTP1B in complex with FMOPL000466a | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tyrosine-protein phosphatase non-receptor type 1, ~{N},~{N},5,6-tetramethylthieno[2,3-d]pyrimidin-4-amine | | Authors: | Mehlman, T, Ginn, H.M, Keedy, D.A. | | Deposit date: | 2024-01-03 | | Release date: | 2024-01-24 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | An expanded view of ligandability in the allosteric enzyme PTP1B from computational reanalysis of large-scale crystallographic data.
Biorxiv, 2024
|
|
7GTZ
 
 | |
2Z7K
 
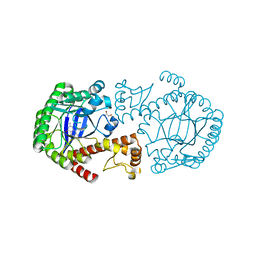 | | tRNA-Guanine transglycosylase (TGT) in complex with 2-Amino-lin-Benzoguanine | | Descriptor: | 2,6-diamino-1,7-dihydro-8H-imidazo[4,5-g]quinazolin-8-one, GLYCEROL, Queuine tRNA-ribosyltransferase, ... | | Authors: | Ritschel, T, Heine, A, Klebe, G. | | Deposit date: | 2007-08-24 | | Release date: | 2008-08-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Crystal structure analysis and in silico pKa calculations suggest strong pKa shifts of ligands as driving force for high-affinity binding to TGT
Chembiochem, 10, 2009
|
|
7GSI
 
 | |
7GU6
 
 | |
7POV
 
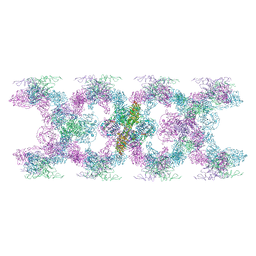 | | MUC2 Tubules of D1D2D3 domains | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Javitt, G, Fass, D. | | Deposit date: | 2021-09-10 | | Release date: | 2022-02-16 | | Last modified: | 2022-05-25 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Helical self-assembly of a mucin segment suggests an evolutionary origin for von Willebrand factor tubules.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7GTC
 
 | |
7GU1
 
 | |
1MQP
 
 | | THE CRYSTAL STRUCTURE OF ALPHA-D-GLUCURONIDASE FROM BACILLUS STEAROTHERMOPHILUS T-6 | | Descriptor: | GLYCEROL, alpha-D-glucuronidase | | Authors: | Golan, G, Shallom, D, Teplitsky, A, Zaide, G, Shulami, S, Baasov, T, Stojanoff, V, Thompson, A, Shoham, Y, Shoham, G. | | Deposit date: | 2002-09-17 | | Release date: | 2003-09-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of Geobacillus stearothermophilus alpha-glucuronidase complexed with its substrate and products: mechanistic implications.
J.Biol.Chem., 279, 2004
|
|
7GTY
 
 | |
3DKW
 
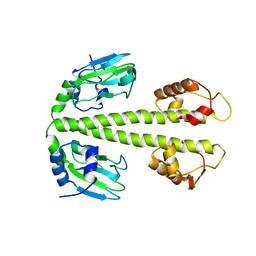 | | Crystal Structure of DNR from Pseudomonas aeruginosa. | | Descriptor: | DNR protein | | Authors: | Giardina, G. | | Deposit date: | 2008-06-26 | | Release date: | 2009-05-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | A dramatic conformational rearrangement is necessary for the activation of DNR from Pseudomonas aeruginosa. Crystal structure of wild-type DNR.
Proteins, 2009
|
|
7GUB
 
 | |
1MX6
 
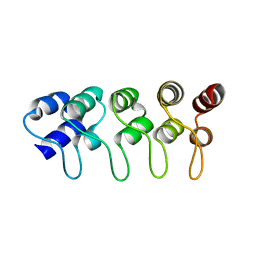 | | Structure of p18INK4c (F92N) | | Descriptor: | Cyclin-dependent kinase 6 inhibitor | | Authors: | Marmorstein, R, Venkataramani, R.N, MacLachlan, T.K, Chai, X, El-Deiry, W.S. | | Deposit date: | 2002-10-01 | | Release date: | 2002-10-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based design of p18INK4c proteins with increased thermodynamic stability and cell cycle inhibitory activity
J.Biol.Chem., 277, 2002
|
|
7GU8
 
 | |
