7ZZ8
 
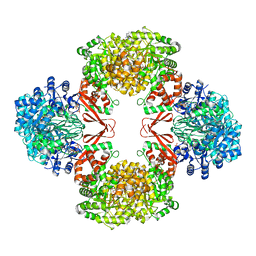 | | Cryo-EM structure of Lactococcus lactis pyruvate carboxylase with acetyl-CoA and cyclic di-AMP | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, ACETYL COENZYME *A, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Lopez-Alonso, J.P, Lazaro, M, Gil, D, Choi, P.H, Tong, L, Valle, M. | | Deposit date: | 2022-05-25 | | Release date: | 2022-10-12 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | CryoEM structural exploration of catalytically active enzyme pyruvate carboxylase.
Nat Commun, 13, 2022
|
|
7ZZ1
 
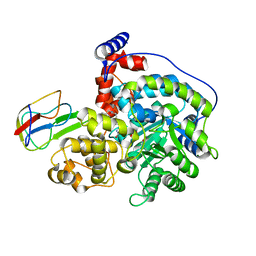 | | Cryo-EM structure of "CT react" conformation of Lactococcus lactis pyruvate carboxylase with acetyl-CoA | | Descriptor: | BIOTIN, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Lopez-Alonso, J.P, Lazaro, M, Gil, D, Choi, P.H, Tong, L, Valle, M. | | Deposit date: | 2022-05-25 | | Release date: | 2022-10-12 | | Last modified: | 2022-11-02 | | Method: | ELECTRON MICROSCOPY (2.27 Å) | | Cite: | CryoEM structural exploration of catalytically active enzyme pyruvate carboxylase.
Nat Commun, 13, 2022
|
|
7ZZ2
 
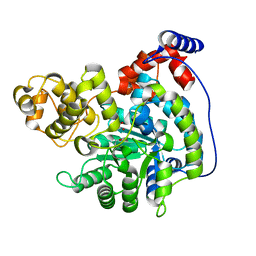 | | Cryo-EM structure of "CT pyr" conformation of Lactococcus lactis pyruvate carboxylase with acetyl-CoA | | Descriptor: | MAGNESIUM ION, MANGANESE (II) ION, PYRUVIC ACID, ... | | Authors: | Lopez-Alonso, J.P, Lazaro, M, Gil, D, Choi, P.H, Tong, L, Valle, M. | | Deposit date: | 2022-05-25 | | Release date: | 2022-10-12 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.48 Å) | | Cite: | CryoEM structural exploration of catalytically active enzyme pyruvate carboxylase.
Nat Commun, 13, 2022
|
|
7ZZ0
 
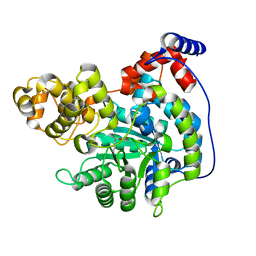 | | Cryo-EM structure of "CT empty" conformation of Lactococcus lactis pyruvate carboxylase with acetyl-CoA | | Descriptor: | MAGNESIUM ION, MANGANESE (II) ION, Pyruvate carboxylase | | Authors: | Lopez-Alonso, J.P, Lazaro, M, Gil, D, Choi, P.H, Tong, L, Valle, M. | | Deposit date: | 2022-05-25 | | Release date: | 2022-10-12 | | Last modified: | 2022-11-02 | | Method: | ELECTRON MICROSCOPY (2.26 Å) | | Cite: | CryoEM structural exploration of catalytically active enzyme pyruvate carboxylase.
Nat Commun, 13, 2022
|
|
7ZYY
 
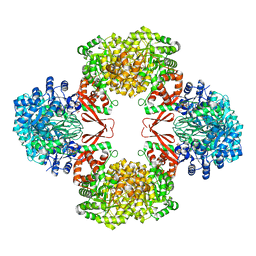 | | Cryo-EM structure of Lactococcus lactis pyruvate carboxylase with acetyl-CoA | | Descriptor: | ACETYL COENZYME *A, ADENOSINE-5'-DIPHOSPHATE, BICARBONATE ION, ... | | Authors: | Lopez-Alonso, J.P, Lazaro, M, Gil, D, Choi, P.H, Tong, L, Valle, M. | | Deposit date: | 2022-05-25 | | Release date: | 2022-10-12 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.12 Å) | | Cite: | CryoEM structural exploration of catalytically active enzyme pyruvate carboxylase.
Nat Commun, 13, 2022
|
|
4HTO
 
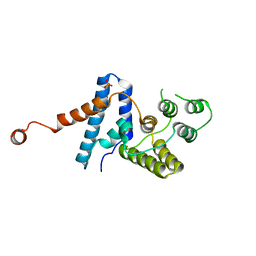 | |
4HTP
 
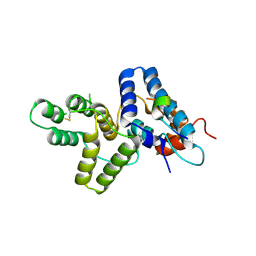 | |
5L6S
 
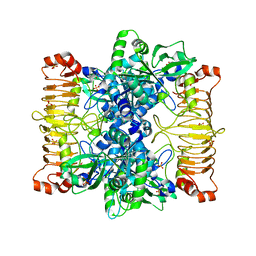 | | Crystal structure of E. coli ADP-glucose pyrophosphorylase (AGPase) in complex with a positive allosteric regulator beta-fructose-1,6-diphosphate (FBP) - AGPase*FBP | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, Glucose-1-phosphate adenylyltransferase, SULFATE ION | | Authors: | Cifuente, J.O, Albesa-Jove, D, Comino, N, Madariaga-Marcos, J, Agirre, J, Lopez-Fernandez, S, Garcia-Alija, M, Guerin, M.E. | | Deposit date: | 2016-05-31 | | Release date: | 2016-09-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.04 Å) | | Cite: | Structural Basis of Glycogen Biosynthesis Regulation in Bacteria.
Structure, 24, 2016
|
|
5KS8
 
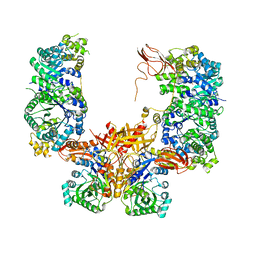 | |
5L6V
 
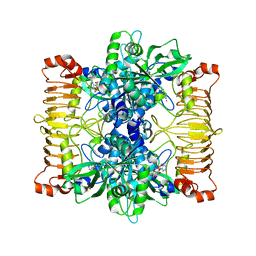 | | Crystal structure of E. coli ADP-glucose pyrophosphorylase (AGPase) in complex with a negative allosteric regulator adenosine monophosphate (AMP) - AGPase*AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Glucose-1-phosphate adenylyltransferase, PHOSPHATE ION, ... | | Authors: | Cifuente, J.O, Albesa-Jove, D, Comino, N, Madariaga-Marcos, J, Agirre, J, Lopez-Fernandez, S, Garcia-Alija, M, Guerin, M.E. | | Deposit date: | 2016-05-31 | | Release date: | 2016-09-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.667 Å) | | Cite: | Structural Basis of Glycogen Biosynthesis Regulation in Bacteria.
Structure, 24, 2016
|
|
6SGA
 
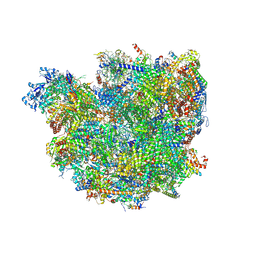 | | Body domain of the mt-SSU assemblosome from Trypanosoma brucei | | Descriptor: | 9S rRNA, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Saurer, M, Ramrath, D.J.F, Niemann, M, Calderaro, S, Prange, C, Mattei, S, Scaiola, A, Leitner, A, Bieri, P, Horn, E.K, Leibundgut, M, Boehringer, D, Schneider, A, Ban, N. | | Deposit date: | 2019-08-03 | | Release date: | 2019-09-18 | | Last modified: | 2019-09-25 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Mitoribosomal small subunit biogenesis in trypanosomes involves an extensive assembly machinery.
Science, 365, 2019
|
|
6SGB
 
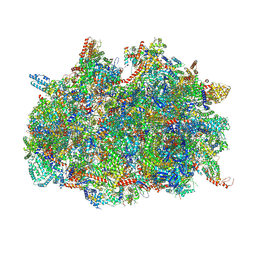 | | mt-SSU assemblosome of Trypanosoma brucei | | Descriptor: | 9S rRNA, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Saurer, M, Ramrath, D.J.F, Niemann, M, Calderaro, S, Prange, C, Mattei, S, Scaiola, A, Leitner, A, Bieri, P, Horn, E.K, Leibundgut, M, Boehringer, D, Schneider, A, Ban, N. | | Deposit date: | 2019-08-03 | | Release date: | 2019-09-25 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Mitoribosomal small subunit biogenesis in trypanosomes involves an extensive assembly machinery.
Science, 365, 2019
|
|
6GQ3
 
 | | Human asparagine synthetase (ASNS) in complex with 6-diazo-5-oxo-L-norleucine (DON) at 1.85 A resolution | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 5-OXO-L-NORLEUCINE, ... | | Authors: | Zhu, W, Radadiya, A, Bisson, C, Jin, Y, Nordin, B.E, Imasaki, T, Wenzel, S, Sedelnikova, S.E, Berry, A.H, Nomanbhoy, T.K, Kozarich, J.W, Takagi, Y, Rice, D.W, Richards, N.G.J. | | Deposit date: | 2018-06-07 | | Release date: | 2019-09-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | High-resolution crystal structure of human asparagine synthetase enables analysis of inhibitor binding and selectivity.
Commun Biol, 2, 2019
|
|
4F4H
 
 | |
2NMM
 
 | | Crystal structure of human phosphohistidine phosphatase. Northeast Structural Genomics Consortium target HR1409 | | Descriptor: | 14 kDa phosphohistidine phosphatase, SULFATE ION | | Authors: | Kuzin, A.P, Abashidze, M, Forouhar, F, Seetharaman, J, Kent, C, Fang, Y, Cunningham, K, Conover, K, Ma, L.C, Xiao, R, Acton, T, Montelione, G, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-10-22 | | Release date: | 2007-01-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of human phosphohistidine phosphatase. Northeast Structural Genomics Consortium target HR1409
To be Published
|
|
4HQ6
 
 | | BC domain in the presence of citrate | | Descriptor: | Acetyl-CoA carboxylase 2 | | Authors: | Heo, Y.S. | | Deposit date: | 2012-10-25 | | Release date: | 2013-10-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Insights into the Regulation of ACC2 by Citrate
Bull.Korean Chem.Soc., 34, 2013
|
|
8J4Z
 
 | |
8J78
 
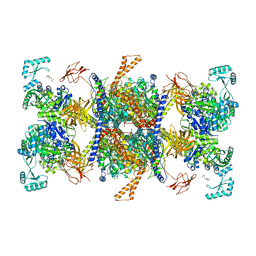 | | Human 3-methylcrotonyl-CoA carboxylase in BCCP-H2 state | | Descriptor: | 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, Methylcrotonoyl-CoA carboxylase beta chain, mitochondrial, ... | | Authors: | Liu, D.S, Su, J.Y. | | Deposit date: | 2023-04-27 | | Release date: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.88 Å) | | Cite: | Human 3-methylcrotonyl-CoA carboxylase in BCCP-H2 state
To Be Published
|
|
8J99
 
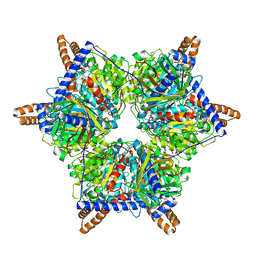 | | Human 3-methylcrotonyl-CoA carboxylase in BCS-mcoa state | | Descriptor: | Methylcrotonoyl-CoA carboxylase beta chain, mitochondrial, Methylcrotonoyl-CoA carboxylase subunit alpha, ... | | Authors: | Liu, D.S, Su, J.Y. | | Deposit date: | 2023-05-02 | | Release date: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Human 3-methylcrotonyl-CoA carboxylase in BCS-mcoa state
To Be Published
|
|
8J7D
 
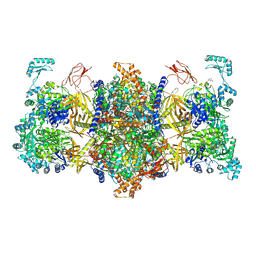 | | Human 3-methylcrotonyl-CoA carboxylase in BCCP-H1 state | | Descriptor: | 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, Methylcrotonoyl-CoA carboxylase beta chain, mitochondrial, ... | | Authors: | Liu, D.S, Su, J.Y. | | Deposit date: | 2023-04-27 | | Release date: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Human 3-methylcrotonyl-CoA carboxylase in BCCP-H1 state
To Be Published
|
|
8JAK
 
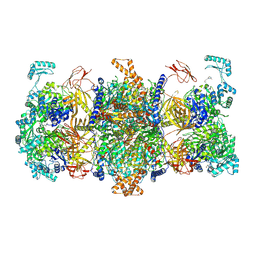 | | Human MCC in MCCU state | | Descriptor: | 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, Methylcrotonoyl-CoA carboxylase beta chain, mitochondrial, ... | | Authors: | Liu, D.S, Su, J.Y, Tian, X.Y. | | Deposit date: | 2023-05-06 | | Release date: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (2.52 Å) | | Cite: | Human MCC in BCCP-BCS state
To Be Published
|
|
8JAW
 
 | | Human MCC in MCCD state | | Descriptor: | 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, Methylcrotonoyl-CoA carboxylase beta chain, mitochondrial, ... | | Authors: | Liu, D.S, Su, J.Y, Tian, X.Y. | | Deposit date: | 2023-05-07 | | Release date: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (2.51 Å) | | Cite: | Human MCC in BCCP-CTS state
To Be Published
|
|
5TV6
 
 | | A. aeolicus BioW with pimelate | | Descriptor: | 6-carboxyhexanoate--CoA ligase, PIMELIC ACID | | Authors: | Estrada, P, Manandhar, M, Dong, S.-H, Deveryshetty, J, Agarwal, V, Cronan, J.E, Nair, S.K. | | Deposit date: | 2016-11-08 | | Release date: | 2016-12-07 | | Last modified: | 2017-05-31 | | Method: | X-RAY DIFFRACTION (2.456 Å) | | Cite: | The pimeloyl-CoA synthetase BioW defines a new fold for adenylate-forming enzymes.
Nat. Chem. Biol., 13, 2017
|
|
5ZK6
 
 | |
5TVA
 
 | | A. aeolicus BioW with AMP and CoA | | Descriptor: | 6-carboxyhexanoate--CoA ligase, ADENOSINE MONOPHOSPHATE, COENZYME A | | Authors: | Estrada, P, Manandhar, M, Dong, S.-H, Deveryshetty, J, Agarwal, V, Cronan, J.E, Nair, S.K. | | Deposit date: | 2016-11-08 | | Release date: | 2016-12-07 | | Last modified: | 2017-05-31 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The pimeloyl-CoA synthetase BioW defines a new fold for adenylate-forming enzymes.
Nat. Chem. Biol., 13, 2017
|
|
