6SCX
 
 | | Crystal structure of the catalytic domain of human NUDT12 in complex with 7-methyl-guanosine-5'-triphosphate | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE, CADMIUM ION, Peroxisomal NADH pyrophosphatase NUDT12 | | Authors: | McCarthy, A.A, Chen, K.M, Wu, H, Li, L, Homolka, D, Gos, P, Fleury-Olela, F, Pillai, R.S. | | Deposit date: | 2019-07-25 | | Release date: | 2020-01-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Decapping Enzyme NUDT12 Partners with BLMH for Cytoplasmic Surveillance of NAD-Capped RNAs.
Cell Rep, 29, 2019
|
|
6O3P
 
 | |
7E44
 
 | | Crystal structure of NudC complexed with dpCoA | | Descriptor: | DEPHOSPHO COENZYME A, NADH pyrophosphatase, ZINC ION | | Authors: | Zhou, W, Guan, Z.Y, Yin, P, Zhang, D.L. | | Deposit date: | 2021-02-10 | | Release date: | 2021-07-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into dpCoA-RNA decapping by NudC.
Rna Biol., 18, 2021
|
|
2FML
 
 | |
2GB5
 
 | |
4HFQ
 
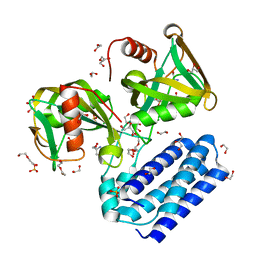 | | Crystal structure of UDP-X diphosphatase | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Duong-Ly, K.C, Amzel, L.M, Gabelli, S.B. | | Deposit date: | 2012-10-05 | | Release date: | 2013-08-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | A UDP-X diphosphatase from Streptococcus pneumoniae hydrolyzes precursors of peptidoglycan biosynthesis.
Plos One, 8, 2013
|
|
2QJT
 
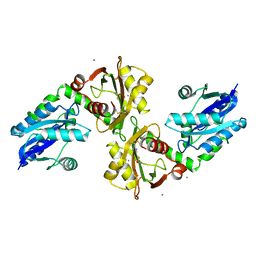 | | Crystal structure of a bifunctional NMN adenylyltransferase/ADP ribose pyrophosphatase complexed with AMP and MN ion from Francisella tularensis | | Descriptor: | ADENOSINE MONOPHOSPHATE, MANGANESE (II) ION, Nicotinamide-nucleotide adenylyltransferase | | Authors: | Huang, N, Sorci, L, Zhang, X, Brautigan, C, Raffaelli, N, Magni, G, Grishin, N.V, Osterman, A, Zhang, H. | | Deposit date: | 2007-07-09 | | Release date: | 2008-03-04 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Bifunctional NMN Adenylyltransferase/ADP-Ribose Pyrophosphatase: Structure and Function in Bacterial NAD Metabolism.
Structure, 16, 2008
|
|
2A6T
 
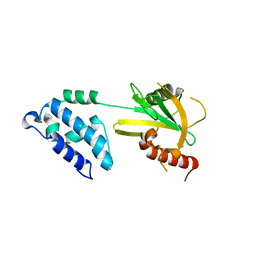 | |
2R5W
 
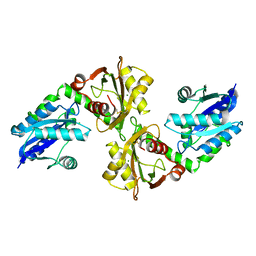 | | Crystal structure of a bifunctional NMN adenylyltransferase/ADP ribose pyrophosphatase from Francisella tularensis | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Nicotinamide-nucleotide adenylyltransferase | | Authors: | Huang, N, Sorci, L, Zhang, X, Brautigan, C, Li, X, Raffaelli, N, Grishin, N, Osterman, A, Zhang, H. | | Deposit date: | 2007-09-04 | | Release date: | 2008-03-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Bifunctional NMN Adenylyltransferase/ADP-Ribose Pyrophosphatase: Structure and Function in Bacterial NAD Metabolism.
Structure, 16, 2008
|
|
3Q4I
 
 | | Crystal structure of CDP-Chase in complex with Gd3+ | | Descriptor: | GADOLINIUM ION, Phosphohydrolase (MutT/nudix family protein) | | Authors: | Duong-Ly, K.C, Gabelli, S.B, Amzel, L.M. | | Deposit date: | 2010-12-23 | | Release date: | 2011-07-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Nudix Hydrolase CDP-Chase, a CDP-Choline Pyrophosphatase, Is an Asymmetric Dimer with Two Distinct Enzymatic Activities.
J.Bacteriol., 193, 2011
|
|
6M65
 
 | | Crystal structure of Mycobacterium smegmatis MutT1 in complex with GMPPNP (GDP) | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Raj, P, Karthik, S, Arif, S.M, Varshney, U, Vijayan, M. | | Deposit date: | 2020-03-13 | | Release date: | 2020-10-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Plasticity, ligand conformation and enzyme action of Mycobacterium smegmatis MutT1.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6M69
 
 | | Crystal structure of Mycobacterium smegmatis MutT1 in complex with GMPPCP (GDP) | | Descriptor: | 1,2-ETHANEDIOL, GUANOSINE-5'-DIPHOSPHATE, Hydrolase, ... | | Authors: | Raj, P, Karthik, S, Arif, S.M, Varshney, U, Vijayan, M. | | Deposit date: | 2020-03-13 | | Release date: | 2020-10-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Plasticity, ligand conformation and enzyme action of Mycobacterium smegmatis MutT1.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6M6Y
 
 | | Crystal structure of Mycobacterium smegmatis MutT1 in complex with 8-oxo-dGTP | | Descriptor: | 1,2-ETHANEDIOL, 8-OXO-2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, Hydrolase, ... | | Authors: | Raj, P, Karthik, S, Arif, S.M, Varshney, U, Vijayan, M. | | Deposit date: | 2020-03-16 | | Release date: | 2020-10-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Plasticity, ligand conformation and enzyme action of Mycobacterium smegmatis MutT1.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6M72
 
 | | Crystal structure of Mycobacterium smegmatis MutT1 in complex with 8-oxo-dGDP | | Descriptor: | 2'-deoxy-8-oxoguanosine 5'-(trihydrogen diphosphate), Hydrolase, NUDIX family protein, ... | | Authors: | Raj, P, Karthik, S, Arif, S.M, Varshney, U, Vijayan, M. | | Deposit date: | 2020-03-16 | | Release date: | 2020-10-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Plasticity, ligand conformation and enzyme action of Mycobacterium smegmatis MutT1.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
5J3T
 
 | | Crystal structure of S. pombe Dcp2:Dcp1:Edc1 mRNA decapping complex | | Descriptor: | Edc1, FORMIC ACID, MAGNESIUM ION, ... | | Authors: | Valkov, E, Muthukumar, S, Chang, C.T, Jonas, S, Weichenrieder, O, Izaurralde, E. | | Deposit date: | 2016-03-31 | | Release date: | 2016-05-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of the Dcp2-Dcp1 mRNA-decapping complex in the activated conformation.
Nat.Struct.Mol.Biol., 23, 2016
|
|
5IW4
 
 | |
6NCI
 
 | | Crystal structure of CDP-Chase: Vector data collection | | Descriptor: | D-ribose, DI(HYDROXYETHYL)ETHER, PHOSPHATE ION, ... | | Authors: | Miller, M.S, Shi, W, Gabelli, S.B. | | Deposit date: | 2018-12-11 | | Release date: | 2019-02-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Getting the Most Out of Your Crystals: Data Collection at the New High-Flux, Microfocus MX Beamlines at NSLS-II.
Molecules, 24, 2019
|
|
5J3Y
 
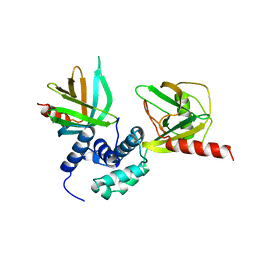 | | Crystal structure of S. pombe Dcp2:Dcp1 mRNA decapping complex | | Descriptor: | mRNA decapping complex subunit 2, mRNA-decapping enzyme subunit 1 | | Authors: | Valkov, E, Muthukumar, S, Chang, C.T, Jonas, S, Weichenrieder, O, Izaurralde, E. | | Deposit date: | 2016-03-31 | | Release date: | 2016-05-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.288 Å) | | Cite: | Structure of the Dcp2-Dcp1 mRNA-decapping complex in the activated conformation.
Nat.Struct.Mol.Biol., 23, 2016
|
|
5ISY
 
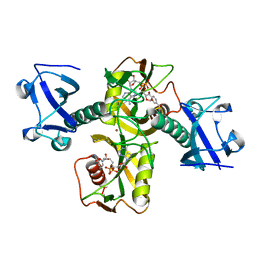 | | Crystal structure of Nudix family protein with NAD | | Descriptor: | NADH pyrophosphatase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ZINC ION | | Authors: | Zhang, D, Guan, Z, Zou, T, Yin, P. | | Deposit date: | 2016-03-15 | | Release date: | 2016-09-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.354 Å) | | Cite: | Structural basis of prokaryotic NAD-RNA decapping by NudC
Cell Res., 26, 2016
|
|
6NCH
 
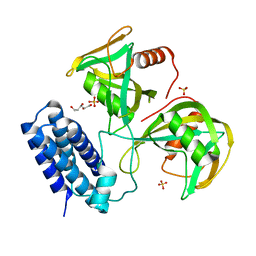 | | Crystal structure of CDP-Chase: Raster data collection | | Descriptor: | D-ribose, PHOSPHATE ION, Phosphohydrolase (MutT/nudix family protein), ... | | Authors: | Miller, M.S, Shi, W, Gabelli, S.B. | | Deposit date: | 2018-12-11 | | Release date: | 2019-02-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Getting the Most Out of Your Crystals: Data Collection at the New High-Flux, Microfocus MX Beamlines at NSLS-II.
Molecules, 24, 2019
|
|
3Q1P
 
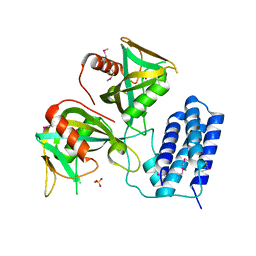 | | Crystal structure of CDP-Chase | | Descriptor: | Phosphohydrolase (MutT/nudix family protein), SULFATE ION | | Authors: | Duong-Ly, K.C, Gabelli, S.B, Amzel, L.M. | | Deposit date: | 2010-12-17 | | Release date: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Nudix Hydrolase CDP-Chase, a CDP-Choline Pyrophosphatase, Is an Asymmetric Dimer with Two Distinct Enzymatic Activities.
J.Bacteriol., 193, 2011
|
|
5IW5
 
 | |
2QKM
 
 | |
2QJO
 
 | | crystal structure of a bifunctional NMN adenylyltransferase/ADP ribose pyrophosphatase (NadM) complexed with ADPRP and NAD from Synechocystis sp. | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Bifunctional NMN adenylyltransferase/Nudix hydrolase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Huang, N, Sorci, L, Zhang, X, Brautigan, C, Raffaelli, N, Magni, G, Grishin, N.V, Osterman, A, Zhang, H. | | Deposit date: | 2007-07-08 | | Release date: | 2008-03-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Bifunctional NMN Adenylyltransferase/ADP-Ribose Pyrophosphatase: Structure and Function in Bacterial NAD Metabolism.
Structure, 16, 2008
|
|
5XD1
 
 | | Crystal structure of Mycobacterium smegmatis MutT1 in complex with Ap5A, ATP and magnesium | | Descriptor: | ADENOSINE-5'-PENTAPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, ... | | Authors: | Arif, S.M, Varshney, U, Vijayan, M. | | Deposit date: | 2017-03-24 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Hydrolysis of diadenosine polyphosphates. Exploration of an additional role of Mycobacterium smegmatis MutT1
J. Struct. Biol., 199, 2017
|
|
