8REE
 
 | |
8EGB
 
 | | Cryo-EM structure of consensus elemental paused elongation complex with an unfolded TL | | Descriptor: | CHAPSO, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Kang, J.Y, Chen, J, Llewellyn, E, Landick, R, Darst, S.A. | | Deposit date: | 2022-09-12 | | Release date: | 2023-03-01 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | An ensemble of interconverting conformations of the elemental paused transcription complex creates regulatory options.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
6ASX
 
 | | CryoEM structure of E.coli his pause elongation complex | | Descriptor: | DNA (32-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Kang, J.Y, Landick, R, Darst, S.A. | | Deposit date: | 2017-08-25 | | Release date: | 2018-03-28 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | RNA Polymerase Accommodates a Pause RNA Hairpin by Global Conformational Rearrangements that Prolong Pausing.
Mol. Cell, 69, 2018
|
|
6UTZ
 
 | | E. coli sigma-S transcription initiation complex with a 6-nt RNA ("Fresh" crystal soaked with CTP and UTP for 30 minutes) | | Descriptor: | DIPHOSPHATE, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Zuo, Y, De, S, Steitz, T.A. | | Deposit date: | 2019-10-30 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.803 Å) | | Cite: | Structural Insights into Transcription Initiation from De Novo RNA Synthesis to Transitioning into Elongation.
Iscience, 23, 2020
|
|
6N62
 
 | | Escherichia coli RNA polymerase sigma70-holoenzyme bound to upstream fork promoter DNA | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Braffman, N, Hauver, J, Campbell, E.A, Darst, S.A. | | Deposit date: | 2018-11-24 | | Release date: | 2019-01-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.803 Å) | | Cite: | Structural mechanism of transcription inhibition by lasso peptides microcin J25 and capistruin.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
5W1S
 
 | |
4KMU
 
 | |
6UTW
 
 | | E. coli sigma-S transcription initiation complex with a 4-nt RNA ("Fresh" crystal) | | Descriptor: | DIPHOSPHATE, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Zuo, Y, De, S, Steitz, T.A. | | Deposit date: | 2019-10-30 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.854 Å) | | Cite: | Structural Insights into Transcription Initiation from De Novo RNA Synthesis to Transitioning into Elongation.
Iscience, 23, 2020
|
|
6PMI
 
 | | Sigm28-transcription initiation complex with specific promoter at the state 1 | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Liu, B, Shi, W. | | Deposit date: | 2019-07-02 | | Release date: | 2020-05-13 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.86 Å) | | Cite: | Structural basis of bacterial sigma28-mediated transcription reveals roles of the RNA polymerase zinc-binding domain.
Embo J., 39, 2020
|
|
5UAL
 
 | | Escherichia coli RNA polymerase and Rifampin complex, RpoB S531L mutant | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Molodtsov, V, Scharf, N.T, Stefan, M.A, Garcia, G.A, Murakami, K.S. | | Deposit date: | 2016-12-19 | | Release date: | 2017-01-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.887 Å) | | Cite: | Structural basis for rifamycin resistance of bacterial RNA polymerase by the three most clinically important RpoB mutations found in Mycobacterium tuberculosis.
Mol. Microbiol., 103, 2017
|
|
6Z9P
 
 | | Transcription termination intermediate complex 1 | | Descriptor: | 2'-DEOXYGUANOSINE-5'-MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Said, N, Hilal, T, Loll, B, Wahl, M.C. | | Deposit date: | 2020-06-04 | | Release date: | 2020-11-04 | | Last modified: | 2021-02-03 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Steps toward translocation-independent RNA polymerase inactivation by terminator ATPase rho.
Science, 371, 2021
|
|
6X2N
 
 | | Mfd-bound E.coli RNA polymerase elongation complex - I state | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA (64-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Llewellyn, E, Chen, J, Kang, J.Y, Darst, S.A. | | Deposit date: | 2020-05-20 | | Release date: | 2021-02-03 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis for transcription complex disruption by the Mfd translocase.
Elife, 10, 2021
|
|
6RI7
 
 | | Cryo-EM structure of E. coli RNA polymerase elongation complex bound to GreB transcription factor | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Abdelkareem, M, Saint-Andre, C, Takacs, M, Papai, G, Crucifix, C, Guo, X, Ortiz, J, Weixlbaumer, A. | | Deposit date: | 2019-04-23 | | Release date: | 2019-07-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural Basis of Transcription: RNA Polymerase Backtracking and Its Reactivation.
Mol.Cell, 75, 2019
|
|
6XH7
 
 | | CueR-TAC without RNA | | Descriptor: | COPPER (II) ION, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Liu, B, Shi, W, Yang, Y. | | Deposit date: | 2020-06-18 | | Release date: | 2021-04-14 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis of copper-efflux-regulator-dependent transcription activation.
Iscience, 24, 2021
|
|
8G7E
 
 | | Cryo-EM structure of 3DVA component 0 of Escherichia coli que-PEC (paused elongation complex) RNA Polymerase plus preQ1 ligand | | Descriptor: | 7-DEAZA-7-AMINOMETHYL-GUANINE, DNA (31-MER), DNA (39-mer), ... | | Authors: | Porta, J.C, Chauvier, A, Deb, I, Ellinger, E, Frank, A.T, Meze, K, Ohi, M.D, Walter, N.G. | | Deposit date: | 2023-02-16 | | Release date: | 2023-06-21 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis for control of bacterial RNA polymerase pausing by a riboswitch and its ligand.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7PY5
 
 | | CryoEM structure of E.coli RNA polymerase elongation complex bound to NusA and NusG (the consensus NusA-NusG-EC) | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Zhu, C, Guo, X, Weixlbaumer, A. | | Deposit date: | 2021-10-09 | | Release date: | 2022-03-23 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Transcription factors modulate RNA polymerase conformational equilibrium.
Nat Commun, 13, 2022
|
|
6B6H
 
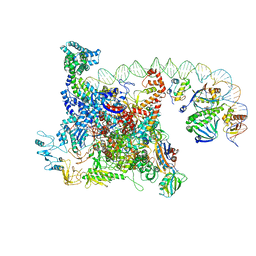 | | The cryo-EM structure of a bacterial class I transcription activation complex | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Liu, B, Hong, C, Huang, R, Yu, Z, Steitz, T.A. | | Deposit date: | 2017-10-02 | | Release date: | 2017-11-15 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis of bacterial transcription activation.
Science, 358, 2017
|
|
6ZTN
 
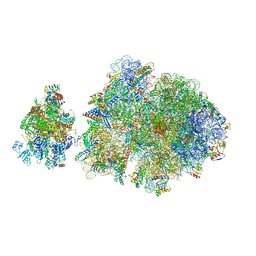 | | E. coli 70S-RNAP expressome complex in NusG-coupled state (42 nt intervening mRNA) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Webster, M.W, Takacs, M, Weixlbaumer, A. | | Deposit date: | 2020-07-20 | | Release date: | 2020-09-16 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis of transcription-translation coupling and collision in bacteria.
Science, 369, 2020
|
|
8FIX
 
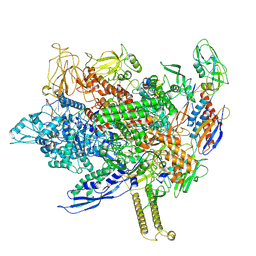 | | Cryo-EM structure of E. coli RNA polymerase backtracked elongation complex harboring a terminal mismatch | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Florez Ariza, A, Wee, L, Tong, A, Canari, C, Grob, P, Nogales, E, Bustamante, C. | | Deposit date: | 2022-12-17 | | Release date: | 2023-03-29 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | A trailing ribosome speeds up RNA polymerase at the expense of transcript fidelity via force and allostery.
Cell, 186, 2023
|
|
8RED
 
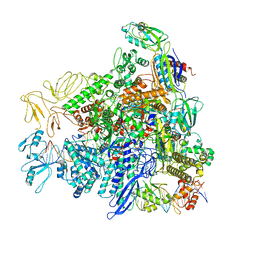 | |
8EH9
 
 | | Cryo-EM structure of his-elemental paused elongation complex with a folded TL and a rotated RH-FL (2) | | Descriptor: | CHAPSO, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Kang, J.Y, Chen, J, Llewellyn, E, Landick, R, Darst, S.A. | | Deposit date: | 2022-09-13 | | Release date: | 2023-03-01 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | An ensemble of interconverting conformations of the elemental paused transcription complex creates regulatory options.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
6PSU
 
 | | Escherichia coli RNA polymerase promoter unwinding intermediate (TRPi2) with TraR and rpsT P2 promoter | | Descriptor: | CHAPSO, DNA (85-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Chen, J, Chiu, C.E, Campbell, E.A, Darst, S.A. | | Deposit date: | 2019-07-13 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Stepwise Promoter Melting by Bacterial RNA Polymerase.
Mol.Cell, 78, 2020
|
|
6UU0
 
 | | E. coli sigma-S transcription initiation complex with a 3-nt RNA and a mismatching GTP ("Fresh" crystal soaked with GTP for 1 hour) | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Zuo, Y, De, S, Steitz, T.A. | | Deposit date: | 2019-10-30 | | Release date: | 2020-08-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Structural Insights into Transcription Initiation from De Novo RNA Synthesis to Transitioning into Elongation.
Iscience, 23, 2020
|
|
4JK1
 
 | |
4MEX
 
 | | Crystal structure of Escherichia coli RNA polymerase in complex with salinamide A | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Feng, Y, Zhang, Y, Arnold, E, Ebright, R.H. | | Deposit date: | 2013-08-27 | | Release date: | 2014-05-21 | | Last modified: | 2014-06-25 | | Method: | X-RAY DIFFRACTION (3.902 Å) | | Cite: | Transcription inhibition by the depsipeptide antibiotic salinamide A.
Elife, 3, 2014
|
|
