1B86
 
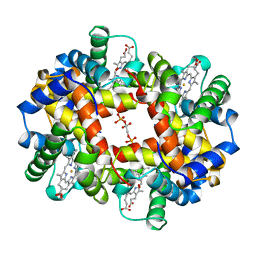 | | HUMAN DEOXYHAEMOGLOBIN-2,3-DIPHOSPHOGLYCERATE COMPLEX | | Descriptor: | (2R)-2,3-diphosphoglyceric acid, OXYGEN MOLECULE, PROTEIN (HEMOGLOBIN; ALPHA CHAIN), ... | | Authors: | Richard, V, Dodson, G.G, Mauguen, Y. | | Deposit date: | 1999-02-08 | | Release date: | 1999-02-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Human deoxyhaemoglobin-2,3-diphosphoglycerate complex low-salt structure at 2.5 A resolution.
J.Mol.Biol., 233, 1993
|
|
1B87
 
 | |
1B88
 
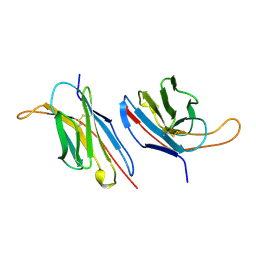 | | V-ALPHA 2.6 MOUSE T CELL RECEPTOR (TCR) DOMAIN | | Descriptor: | T CELL RECEPTOR V-ALPHA DOMAIN | | Authors: | Plaksin, D, Chacko, S, Navaza, J, Margulies, D.H, Padlan, E.A. | | Deposit date: | 1999-02-09 | | Release date: | 1999-02-16 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The X-ray crystal structure of a Valpha2.6Jalpha38 mouse T cell receptor domain at 2.5 A resolution: alternate modes of dimerization and crystal packing.
J.Mol.Biol., 289, 1999
|
|
1B89
 
 | | CLATHRIN HEAVY CHAIN PROXIMAL LEG SEGMENT (BOVINE) | | Descriptor: | PROTEIN (CLATHRIN HEAVY CHAIN) | | Authors: | Ybe, J.A, Brodsky, F.M, Hofmann, K, Lin, K, Liu, S.-H, Chen, L, Earnest, T.N, Fletterick, R.J, Hwang, P.K. | | Deposit date: | 1999-05-27 | | Release date: | 1999-06-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Clathrin self-assembly is mediated by a tandemly repeated superhelix.
Nature, 399, 1999
|
|
1B8A
 
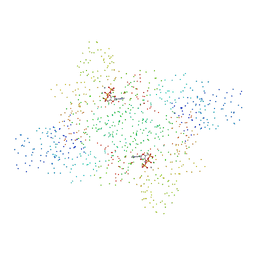 | | ASPARTYL-TRNA SYNTHETASE | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, PROTEIN (ASPARTYL-TRNA SYNTHETASE) | | Authors: | Schmitt, E, Moulinier, L, Thierry, J.-C, Moras, D. | | Deposit date: | 1999-01-27 | | Release date: | 1999-02-02 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of aspartyl-tRNA synthetase from Pyrococcus kodakaraensis KOD: archaeon specificity and catalytic mechanism of adenylate formation.
EMBO J., 17, 1998
|
|
1B8C
 
 | | PARVALBUMIN | | Descriptor: | MAGNESIUM ION, PROTEIN (PARVALBUMIN) | | Authors: | Cates, M.S, Berry, M.B, Ho, E.L, Li, Q, Potter, J.D, Phillips Jr, G.N. | | Deposit date: | 1999-01-29 | | Release date: | 1999-02-02 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Metal-ion affinity and specificity in EF-hand proteins: coordination geometry and domain plasticity in parvalbumin.
Structure Fold.Des., 7, 1999
|
|
1B8D
 
 | | CRYSTAL STRUCTURE OF A PHYCOUROBILIN-CONTAINING PHYCOERYTHRIN | | Descriptor: | PHYCOERYTHROBILIN, PHYCOUROBILIN, PROTEIN (RHODOPHYTAN PHYCOERYTHRIN (ALPHA CHAIN)), ... | | Authors: | Ritter, S, Hiller, R.G, Wrench, P.M, Welte, W, Diederichs, K. | | Deposit date: | 1999-01-29 | | Release date: | 1999-02-18 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a phycourobilin-containing phycoerythrin at 1.90-A resolution.
J.Struct.Biol., 126, 1999
|
|
1B8E
 
 | |
1B8F
 
 | | Histidine ammonia-lyase (HAL) from Pseudomonas putida | | Descriptor: | GLYCEROL, Histidine ammonia-lyase, SULFATE ION | | Authors: | Schwede, T.F, Schulz, G.E. | | Deposit date: | 1999-01-31 | | Release date: | 1999-05-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of histidine ammonia-lyase revealing a novel polypeptide modification as the catalytic electrophile.
Biochemistry, 38, 1999
|
|
1B8G
 
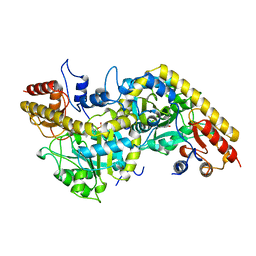 | | 1-AMINOCYCLOPROPANE-1-CARBOXYLATE SYNTHASE | | Descriptor: | PROTEIN (1-AMINOCYCLOPROPANE-1-CARBOXYLATE SYNTHASE), PYRIDOXAL-5'-PHOSPHATE | | Authors: | Capitani, G, Hohenester, E, Feng, L, Storici, P, Kirsch, J.F, Jansonius, J.N. | | Deposit date: | 1999-01-31 | | Release date: | 2000-01-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structure of 1-aminocyclopropane-1-carboxylate synthase, a key enzyme in the biosynthesis of the plant hormone ethylene.
J.Mol.Biol., 294, 1999
|
|
1B8H
 
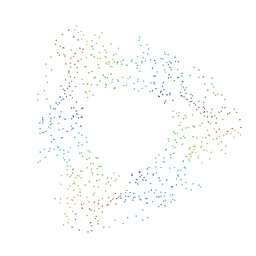 | | SLIDING CLAMP, DNA POLYMERASE | | Descriptor: | DNA POLYMERASE PROCESSIVITY COMPONENT, DNA POLYMERASE fragment | | Authors: | Shamoo, Y, Steitz, T.A. | | Deposit date: | 1999-02-01 | | Release date: | 1999-02-09 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Building a replisome from interacting pieces: sliding clamp complexed to a peptide from DNA polymerase and a polymerase editing complex.
Cell(Cambridge,Mass.), 99, 1999
|
|
1B8I
 
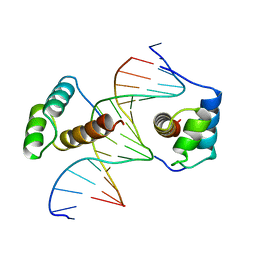 | | STRUCTURE OF THE HOMEOTIC UBX/EXD/DNA TERNARY COMPLEX | | Descriptor: | DNA (5'-D(*AP*CP*GP*TP*GP*AP*TP*TP*TP*AP*TP*GP*GP*CP*G)-3'), DNA (5'-D(*GP*TP*CP*GP*CP*CP*AP*TP*AP*AP*AP*TP*CP*AP*C)-3'), PROTEIN (HOMEOBOX PROTEIN EXTRADENTICLE), ... | | Authors: | Passner, J.M, Ryoo, H.-D, Shen, L, Mann, R.S, Aggarwal, A.K. | | Deposit date: | 1999-02-01 | | Release date: | 1999-04-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of a DNA-bound Ultrabithorax-Extradenticle homeodomain complex.
Nature, 397, 1999
|
|
1B8J
 
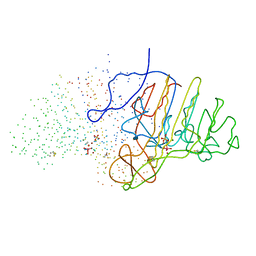 | | ALKALINE PHOSPHATASE COMPLEXED WITH VANADATE | | Descriptor: | MAGNESIUM ION, PROTEIN (ALKALINE PHOSPHATASE), SULFATE ION, ... | | Authors: | Holtz, K.M, Stec, B, Kantrowitz, E.R. | | Deposit date: | 1999-02-01 | | Release date: | 1999-02-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A model of the transition state in the alkaline phosphatase reaction.
J.Biol.Chem., 274, 1999
|
|
1B8K
 
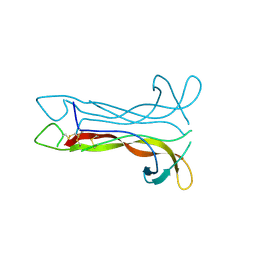 | | Neurotrophin-3 from Human | | Descriptor: | PROTEIN (NEUROTROPHIN-3) | | Authors: | Robinson, R.C, Radziejewski, C, Stuart, D.I, Jones, E.Y, Choe, S. | | Deposit date: | 1999-02-01 | | Release date: | 1999-02-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The structures of the neurotrophin 4 homodimer and the brain-derived neurotrophic factor/neurotrophin 4 heterodimer reveal a common Trk-binding site.
Protein Sci., 8, 1999
|
|
1B8L
 
 | | Calcium-bound D51A/E101D/F102W Triple Mutant of Beta Carp Parvalbumin | | Descriptor: | CALCIUM ION, CARBONATE ION, PROTEIN (PARVALBUMIN) | | Authors: | Cates, M.S, Berry, M.B, Ho, E, Li, Q, Potter, J.D, Phillips Jr, G.N. | | Deposit date: | 1999-02-01 | | Release date: | 1999-10-05 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Metal-ion affinity and specificity in EF-hand proteins: coordination geometry and domain plasticity in parvalbumin.
Structure Fold.Des., 7, 1999
|
|
1B8M
 
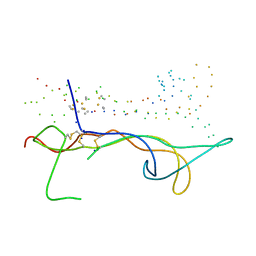 | | BRAIN DERIVED NEUROTROPHIC FACTOR, NEUROTROPHIN-4 | | Descriptor: | PROTEIN (BRAIN DERIVED NEUROTROPHIC FACTOR), PROTEIN (NEUROTROPHIN-4) | | Authors: | Robinson, R.C, Radziejewski, C, Stuart, D.I, Jones, E.Y, Choe, S. | | Deposit date: | 1999-02-01 | | Release date: | 1999-02-09 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The structures of the neurotrophin 4 homodimer and the brain-derived neurotrophic factor/neurotrophin 4 heterodimer reveal a common Trk-binding site.
Protein Sci., 8, 1999
|
|
1B8N
 
 | | PURINE NUCLEOSIDE PHOSPHORYLASE | | Descriptor: | 1,4-DIDEOXY-1,4-IMINO-1-(S)-(9-DEAZAGUANIN-9-YL)-D-RIBITOL, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Fedorov, A.A, Kicska, G.A, Fedorov, E.V, Strokopytov, B.V, Tyler, P.C, Furneaux, R.H, Schramm, V.L, Almo, S.C. | | Deposit date: | 1999-02-02 | | Release date: | 1999-02-08 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Atomic dissection of the hydrogen bond network for transition-state analogue binding to purine nucleoside phosphorylase
Biochemistry, 41, 2002
|
|
1B8O
 
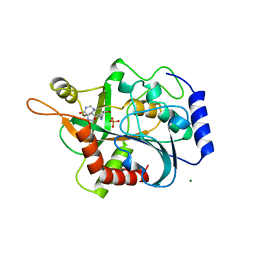 | | PURINE NUCLEOSIDE PHOSPHORYLASE | | Descriptor: | 1,4-DIDEOXY-4-AZA-1-(S)-(9-DEAZAHYPOXANTHIN-9-YL)-D-RIBITOL, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Fedorov, A.A, Kicska, G.A, Fedorov, E.V, Shi, W, Tyler, P.C, Furneaux, R.H, Schramm, V.L, Almo, S.C. | | Deposit date: | 1999-02-02 | | Release date: | 1999-02-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Transition state structure of purine nucleoside phosphorylase and principles of atomic motion in enzymatic catalysis.
Biochemistry, 40, 2001
|
|
1B8P
 
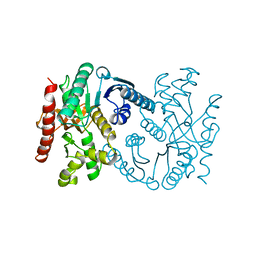 | | MALATE DEHYDROGENASE FROM AQUASPIRILLUM ARCTICUM | | Descriptor: | PROTEIN (MALATE DEHYDROGENASE) | | Authors: | Kim, S.Y, Hwang, K.Y, Kim, S.-H, Han, Y.S, Cho, Y. | | Deposit date: | 1999-02-02 | | Release date: | 1999-07-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for cold adaptation. Sequence, biochemical properties, and crystal structure of malate dehydrogenase from a psychrophile Aquaspirillium arcticum.
J.Biol.Chem., 274, 1999
|
|
1B8Q
 
 | | SOLUTION STRUCTURE OF THE EXTENDED NEURONAL NITRIC OXIDE SYNTHASE PDZ DOMAIN COMPLEXED WITH AN ASSOCIATED PEPTIDE | | Descriptor: | PROTEIN (HEPTAPEPTIDE), PROTEIN (NEURONAL NITRIC OXIDE SYNTHASE) | | Authors: | Tochio, H, Zhang, Q, Mandal, P, Li, M, Zhang, M. | | Deposit date: | 1999-02-01 | | Release date: | 1999-04-29 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the extended neuronal nitric oxide synthase PDZ domain complexed with an associated peptide.
Nat.Struct.Biol., 6, 1999
|
|
1B8R
 
 | | PARVALBUMIN | | Descriptor: | CALCIUM ION, PROTEIN (PARVALBUMIN) | | Authors: | Cates, M.S, Berry, M.B, Ho, E.L, Li, Q, Potter, J.D, Phillips Jr, G.N. | | Deposit date: | 1999-02-02 | | Release date: | 1999-02-09 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Metal-ion affinity and specificity in EF-hand proteins: coordination geometry and domain plasticity in parvalbumin.
Structure Fold.Des., 7, 1999
|
|
1B8S
 
 | | CHOLESTEROL OXIDASE FROM STREPTOMYCES GLU361GLN MUTANT | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, PROTEIN (CHOLESTEROL OXIDASE) | | Authors: | Vrielink, A, Yue, Q.K. | | Deposit date: | 1999-02-02 | | Release date: | 1999-02-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure determination of cholesterol oxidase from Streptomyces and structural characterization of key active site mutants.
Biochemistry, 38, 1999
|
|
1B8T
 
 | | SOLUTION STRUCTURE OF THE CHICKEN CRP1 | | Descriptor: | PROTEIN (CRP1), ZINC ION | | Authors: | Yao, X, Perez-Alvarado, G.C, Louis, H.A, Pomies, P, Hatt, C, Summers, M.F, Beckerle, M.C. | | Deposit date: | 1999-02-02 | | Release date: | 1999-05-06 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the chicken cysteine-rich protein, CRP1, a double-LIM protein implicated in muscle differentiation.
Biochemistry, 38, 1999
|
|
1B8U
 
 | | MALATE DEHYDROGENASE FROM AQUASPIRILLUM ARCTICUM | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, OXALOACETATE ION, PROTEIN (MALATE DEHYDROGENASE) | | Authors: | Kim, S.Y, Hwang, K.Y, Kim, S.-H, Han, Y.S, Cho, Y. | | Deposit date: | 1999-02-02 | | Release date: | 1999-07-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for cold adaptation. Sequence, biochemical properties, and crystal structure of malate dehydrogenase from a psychrophile Aquaspirillium arcticum.
J.Biol.Chem., 274, 1999
|
|
1B8V
 
 | | Malate dehydrogenase from Aquaspirillum arcticum | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PROTEIN (MALATE DEHYDROGENASE) | | Authors: | Kim, S.Y, Hwang, K.Y, Kim, S.H, Han, Y.S, Cho, Y. | | Deposit date: | 1999-02-02 | | Release date: | 1999-07-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for cold adaptation. Sequence, biochemical properties, and crystal structure of malate dehydrogenase from a psychrophile Aquaspirillium arcticum.
J.Biol.Chem., 274, 1999
|
|
