1KXU
 
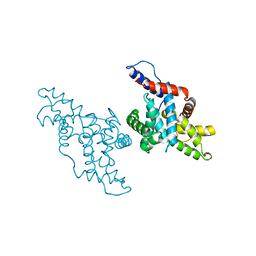 | | CYCLIN H, A POSITIVE REGULATORY SUBUNIT OF CDK ACTIVATING KINASE | | Descriptor: | CYCLIN H | | Authors: | Kim, K.K, Chamberin, H.M, Morgan, D.O, Kim, S.-H. | | Deposit date: | 1996-08-08 | | Release date: | 1997-01-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Three-dimensional structure of human cyclin H, a positive regulator of the CDK-activating kinase.
Nat.Struct.Biol., 3, 1996
|
|
1KXV
 
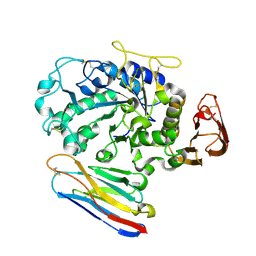 | | Camelid VHH Domains in Complex with Porcine Pancreatic alpha-Amylase | | Descriptor: | ALPHA-AMYLASE, PANCREATIC, CAMELID VHH DOMAIN CAB10 | | Authors: | Desmyter, A, Spinelli, S, Payan, F, Lauwereys, M, Wyns, L, Muyldermans, S, Cambillau, C. | | Deposit date: | 2002-02-01 | | Release date: | 2002-06-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Three camelid VHH domains in complex with porcine pancreatic alpha-amylase. Inhibition and versatility of binding topology.
J.Biol.Chem., 277, 2002
|
|
1KXW
 
 | |
1KXX
 
 | |
1KXY
 
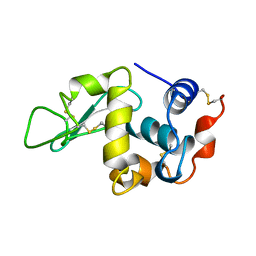 | |
1KXZ
 
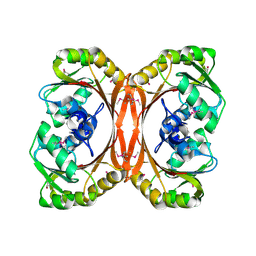 | | MT0146, the Precorrin-6y methyltransferase (CbiT) homolog from M. Thermoautotrophicum, P1 spacegroup | | Descriptor: | Precorrin-6y methyltransferase/putative decarboxylase | | Authors: | Keller, J.P, Smith, P.M, Benach, J, Christendat, D, DeTitta, G, Hunt, J.F. | | Deposit date: | 2002-02-01 | | Release date: | 2002-11-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Crystal Structure of MT0146/CbiT Suggests that the Putative
Precorrin-8W Decarboxylase is a Methyltransferase
Structure, 10, 2002
|
|
1KY0
 
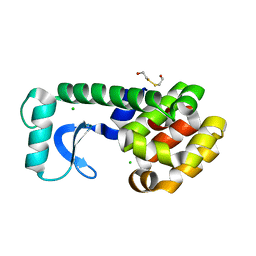 | | METHIONINE CORE MUTANT OF T4 LYSOZYME | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, LYSOZYME | | Authors: | Gassner, N.C, Baase, W.A, Mooers, B.H, Busam, R.D, Weaver, L.H, Lindstrom, J.D, Quillin, M.L, Matthews, B.W. | | Deposit date: | 2002-02-01 | | Release date: | 2003-06-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Multiple methionine substitutions are tolerated in T4 lysozyme and have coupled effects on folding and stability
BIOPHYS.CHEM., 100, 2003
|
|
1KY1
 
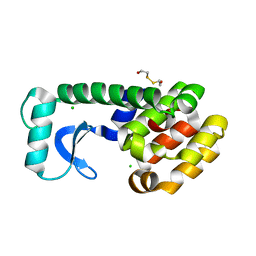 | | METHIONINE CORE MUTANT OF T4 LYSOZYME | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, LYSOZYME | | Authors: | Gassner, N.C, Baase, W.A, Mooers, B.H, Busam, R.D, Weaver, L.H, Lindstrom, J.D, Quillin, M.L, Matthews, B.W. | | Deposit date: | 2002-02-01 | | Release date: | 2003-06-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Multiple methionine substitutions are tolerated in T4 lysozyme and have coupled effects on folding and stability
BIOPHYS.CHEM., 100, 2003
|
|
1KY2
 
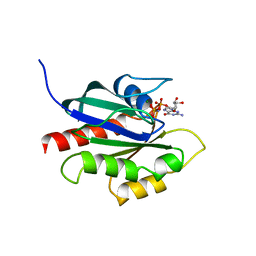 | | GPPNHP-BOUND YPT7P AT 1.6 A RESOLUTION | | Descriptor: | GTP-BINDING PROTEIN YPT7P, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Constantinescu, A.-T, Rak, A, Scheidig, A.J. | | Deposit date: | 2002-02-02 | | Release date: | 2002-06-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Rab-subfamily-specific regions of Ypt7p are structurally different from other RabGTPases.
Structure, 10, 2002
|
|
1KY3
 
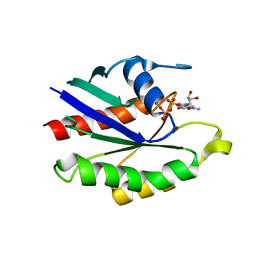 | | GDP-BOUND YPT7P AT 1.35 A RESOLUTION | | Descriptor: | GTP-BINDING PROTEIN YPT7P, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Constantinescu, A.-T, Rak, A, Scheidig, A.J. | | Deposit date: | 2002-02-02 | | Release date: | 2002-06-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Rab-subfamily-specific regions of Ypt7p are structurally different from other RabGTPases.
Structure, 10, 2002
|
|
1KY4
 
 | |
1KY5
 
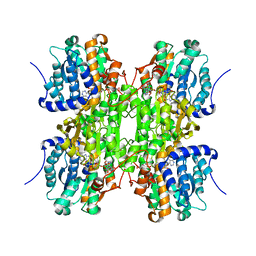 | | D244E mutant S-Adenosylhomocysteine hydrolase refined with noncrystallographic restraints | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 3'-OXO-ADENOSINE, S-adenosylhomocysteine hydrolase | | Authors: | Takata, Y, Takusagawa, F. | | Deposit date: | 2002-02-03 | | Release date: | 2002-09-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Catalytic Mechanism of S-adenosylhomocysteine hydrolase. Site-directed
mutagenesis of Asp-130, Lys-185, Asp-189, and Asn-190.
J.Biol.Chem., 277, 2002
|
|
1KY6
 
 | |
1KY7
 
 | |
1KY8
 
 | | Crystal Structure of the Non-phosphorylating glyceraldehyde-3-phosphate Dehydrogenase | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SODIUM ION, glyceraldehyde-3-phosphate dehydrogenase | | Authors: | Pohl, E, Brunner, N, Wilmanns, M, Hensel, R. | | Deposit date: | 2002-02-04 | | Release date: | 2003-02-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Crystal Structure of the Allosteric Non-phosphorylating
glyceraldehyde-3-phosphate Dehydrogenase from the Hyperthermophilic
Archaeum Thermoproteus tenax
J.Biol.Chem., 277, 2002
|
|
1KY9
 
 | | Crystal Structure of DegP (HtrA) | | Descriptor: | PROTEASE DO | | Authors: | Krojer, T, Garrido-Franco, M, Huber, R, Ehrmann, M, Clausen, T. | | Deposit date: | 2002-02-04 | | Release date: | 2002-04-03 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of DegP (HtrA) reveals a new protease-chaperone machine.
Nature, 416, 2002
|
|
1KYA
 
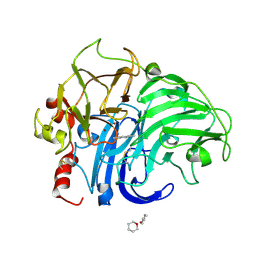 | | ACTIVE LACCASE FROM TRAMETES VERSICOLOR COMPLEXED WITH 2,5-XYLIDINE | | Descriptor: | 2,5-DIMETHYLANILINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, ... | | Authors: | Bertrand, T, Jolivalt, C, Briozzo, P, Caminade, E, Joly, N, Madzak, C, Mougin, C. | | Deposit date: | 2002-02-04 | | Release date: | 2002-06-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of a four-copper laccase complexed with an arylamine: insights into substrate recognition and correlation with kinetics.
Biochemistry, 41, 2002
|
|
1KYC
 
 | |
1KYD
 
 | |
1KYF
 
 | |
1KYH
 
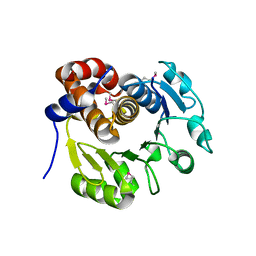 | | Structure of Bacillus subtilis YxkO, a Member of the UPF0031 Family and a Putative Kinase | | Descriptor: | Hypothetical 29.9 kDa protein in SIGY-CYDD intergenic region | | Authors: | Zhang, R, Dementieva, I, Vinokour, E, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-02-04 | | Release date: | 2002-08-14 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of Bacillus subtilis YXKO--a member of the UPF0031 family and a putative kinase.
J.Struct.Biol., 139, 2002
|
|
1KYI
 
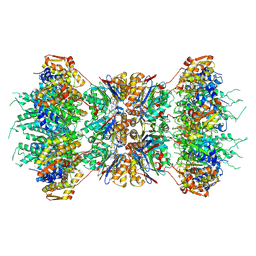 | | HslUV (H. influenzae)-NLVS Vinyl Sulfone Inhibitor Complex | | Descriptor: | 4-IODO-3-NITROPHENYL ACETYL-LEUCINYL-LEUCINYL-LEUCINYL-VINYLSULFONE, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent hsl protease ATP-binding subunit hslU, ... | | Authors: | Sousa, M.C, Kessler, B.M, Overkleeft, H.S, McKay, D.B. | | Deposit date: | 2002-02-04 | | Release date: | 2002-05-15 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal Structure of HslUV Complexed with a Vinyl Sulfone Inhibitor:
Corroboration of a Proposed Mechanism of Allosteric Activation of
HslV by HslU
J.Mol.Biol., 318, 2002
|
|
1KYJ
 
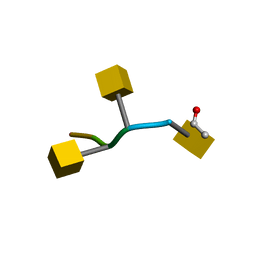 | | Tumor Associated Mucin Motif from CD43 protein | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, Leukosialin (CD43) fragment | | Authors: | Coltart, D.M, Williams, L.J, Glunz, P.W, Sames, D, Kuduk, S.D, Schwarz, J.B, Chen, X.-T, Royyuru, A.K, Danishefsky, S.D, Live, D.H. | | Deposit date: | 2002-02-04 | | Release date: | 2002-02-20 | | Last modified: | 2020-07-29 | | Method: | SOLUTION NMR | | Cite: | Principles of Mucin Architecture: Structural Studies on Synthetic Glycopeptides Bearing Clustered Mono-, Di-, Tri-, and Hexasaccharide Glycodomains
J.Am.Chem.Soc., 124, 2002
|
|
1KYN
 
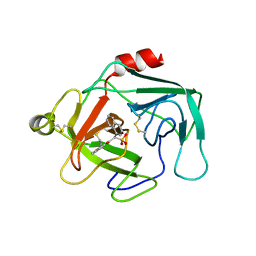 | | Cathepsin-G | | Descriptor: | (2-NAPHTHALEN-2-YL-1-NAPHTHALEN-1-YL-2-OXO-ETHYL)-PHOSPHONIC ACID, cathepsin G | | Authors: | Greco, M.N, Hawkins, M.J, Powell, E.T, Almond Jr, H.R, Corcoran, T.W, De Garavilla, L, Kauffman, J.A, Recacha, R, Chattopadhyay, D, Andrade-Gordon, P, Maryanoff, B.E. | | Deposit date: | 2002-02-05 | | Release date: | 2002-05-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Nonpeptide inhibitors of cathepsin G: optimization of a novel beta-ketophosphonic acid lead by structure-based drug design.
J.Am.Chem.Soc., 124, 2002
|
|
1KYO
 
 | |
