1IXM
 
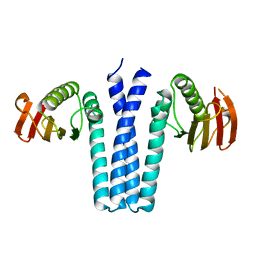 | | CRYSTAL STRUCTURE OF SPOOB FROM BACILLUS SUBTILIS | | Descriptor: | PROTEIN (SPORULATION RESPONSE REGULATORY PROTEIN) | | Authors: | Varughese, K.I. | | Deposit date: | 1998-11-06 | | Release date: | 1998-11-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Formation of a novel four-helix bundle and molecular recognition sites by dimerization of a response regulator phosphotransferase.
Mol.Cell, 2, 1998
|
|
1IXN
 
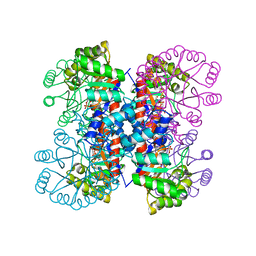 | | Enzyme-Substrate Complex of Pyridoxine 5'-Phosphate Synthase | | Descriptor: | 1-DEOXY-D-XYLULOSE-5-PHOSPHATE, Pyridoxine 5'-Phosphate Synthase, SN-GLYCEROL-3-PHOSPHATE | | Authors: | Garrido-Franco, M, Laber, B, Huber, R, Clausen, T. | | Deposit date: | 2002-06-28 | | Release date: | 2003-02-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Enzyme-ligand complexes of pyridoxine 5'-phosphate synthase: implications for substrate binding and catalysis
J.MOL.BIOL., 321, 2002
|
|
1IXO
 
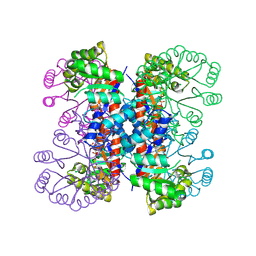 | | Enzyme-analogue substrate complex of Pyridoxine 5'-Phosphate Synthase | | Descriptor: | Pyridoxine 5'-Phosphate synthase, SN-GLYCEROL-3-PHOSPHATE | | Authors: | Garrido-Franco, M, Laber, B, Huber, R, Clausen, T. | | Deposit date: | 2002-06-28 | | Release date: | 2003-02-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Enzyme-ligand complexes of pyridoxine 5'-phosphate synthase: implications for substrate binding and catalysis
J.MOL.BIOL., 321, 2002
|
|
1IXP
 
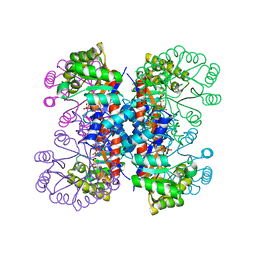 | | Enzyme-phosphate Complex of Pyridoxine 5'-Phosphate synthase | | Descriptor: | PHOSPHATE ION, Pyridoxine 5'-Phosphate synthase | | Authors: | Garrido-Franco, M, Laber, B, Huber, R, Clausen, T. | | Deposit date: | 2002-06-28 | | Release date: | 2003-02-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Enzyme-ligand complexes of pyridoxine 5'-phosphate synthase: implications for substrate binding and catalysis
J.MOL.BIOL., 321, 2002
|
|
1IXQ
 
 | | Enzyme-Phosphate2 Complex of Pyridoxine 5'-Phosphate synthase | | Descriptor: | PHOSPHATE ION, Pyridoxine 5'-phosphate Synthase | | Authors: | Garrido-Franco, M, Laber, B, Huber, R, Clausen, T. | | Deposit date: | 2002-06-28 | | Release date: | 2003-02-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Enzyme-ligand complexes of pyridoxine 5'-phosphate synthase: implications for substrate binding and catalysis
J.MOL.BIOL., 321, 2002
|
|
1IXR
 
 | | RuvA-RuvB complex | | Descriptor: | Holliday junction DNA helicase ruvA, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, RuvB | | Authors: | Yamada, K, Miyata, T, Tsuchiya, D, Oyama, T, Fujiwara, Y, Ohnishi, T, Iwasaki, H, Shinagawa, H, Ariyoshi, M, Mayanagi, K, Morikawa, K. | | Deposit date: | 2002-07-04 | | Release date: | 2002-11-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal Structure of the RuvA-RuvB Complex: A Structural Basis for the Holliday Junction Migrating Motor Machinery
Mol.Cell, 10, 2002
|
|
1IXS
 
 | | Structure of RuvB complexed with RuvA domain III | | Descriptor: | Holliday junction DNA helicase ruvA, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, RuvB | | Authors: | Yamada, K, Miyata, T, Tsuchiya, D, Oyama, T, Fujiwara, Y, Ohnishi, T, Iwasaki, H, Shinagawa, H, Ariyoshi, M, Mayanagi, K, Morikawa, K. | | Deposit date: | 2002-07-04 | | Release date: | 2002-11-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal Structure of the RuvA-RuvB Complex: A Structural Basis for the Holliday Junction Migrating Motor Machinery
Mol.Cell, 10, 2002
|
|
1IXT
 
 | | Structure of a Novel P-Superfamily Spasmodic Conotoxin Reveals an Inhibitory Cystine Knot Motif | | Descriptor: | spasmodic protein tx9a-like protein | | Authors: | Miles, L.A, Dy, C.Y, Nielsen, J, Barnham, K.J, Hinds, M.G, Olivera, B.M, Bulaj, G, Norton, R.S. | | Deposit date: | 2002-07-04 | | Release date: | 2003-01-28 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure of a Novel P-Superfamily Spasmodic Conotoxin Reveals an Inhibitory Cystine Knot Motif
J.Biol.Chem., 277, 2002
|
|
1IXU
 
 | | Solution structure of marinostatin, a protease inhibitor, containing two ester linkages | | Descriptor: | marinostatin | | Authors: | Kanaori, K, Kamei, K, Koyama, T, Yasui, T, Takano, R, Imada, C, Tajima, K, Hara, S. | | Deposit date: | 2002-07-04 | | Release date: | 2004-02-17 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of marinostatin, a natural ester-linked protein protease inhibitor
Biochemistry, 44, 2005
|
|
1IXV
 
 | | Crystal Structure Analysis of homolog of oncoprotein gankyrin, an interactor of Rb and CDK4/6 | | Descriptor: | Probable 26S proteasome regulatory subunit p28 | | Authors: | Padmanabhan, B, Adachi, N, Kataoka, K, Horikoshi, M. | | Deposit date: | 2002-07-09 | | Release date: | 2003-12-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the homolog of the oncoprotein gankyrin, an interactor of Rb and CDK4/6
J.BIOL.CHEM., 279, 2004
|
|
1IXX
 
 | | CRYSTAL STRUCTURE OF COAGULATION FACTORS IX/X-BINDING PROTEIN (IX/X-BP) FROM VENOM OF HABU SNAKE WITH A HETERODIMER OF C-TYPE LECTIN DOMAINS | | Descriptor: | CALCIUM ION, COAGULATION FACTORS IX/X-BINDING PROTEIN | | Authors: | Mizuno, H, Fujimoto, Z, Koizumi, M, Kano, H. | | Deposit date: | 1997-05-01 | | Release date: | 1998-05-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of coagulation factors IX/X-binding protein, a heterodimer of C-type lectin domains.
Nat.Struct.Biol., 4, 1997
|
|
1IXY
 
 | | Ternary complex of T4 phage BGT with UDP and a 13 mer DNA duplex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 5'-D(*CP*TP*AP*TP*CP*TP*GP*AP*GP*TP*AP*TP*C)-3', 5'-D(*GP*AP*TP*AP*CP*TP*3DRP*AP*GP*AP*TP*AP*G)-3', ... | | Authors: | Lariviere, L, Morera, S. | | Deposit date: | 2002-07-09 | | Release date: | 2002-12-04 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A Base-flipping Mechanism for the T4 Phage beta-Glucosyltransferase and Identification of a
Transition-state Analog
J.Mol.Biol., 324, 2002
|
|
1IXZ
 
 | | Crystal structure of the FtsH ATPase domain from Thermus thermophilus | | Descriptor: | ATP-dependent metalloprotease FtsH, MERCURY (II) ION, SULFATE ION | | Authors: | Niwa, H, Tsuchiya, D, Makyio, H, Yoshida, M, Morikawa, K. | | Deposit date: | 2002-07-10 | | Release date: | 2002-11-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Hexameric ring structure of the ATPase domain of the membrane-integrated metalloprotease FtsH from Thermus thermophilus HB8
Structure, 10, 2002
|
|
1IY0
 
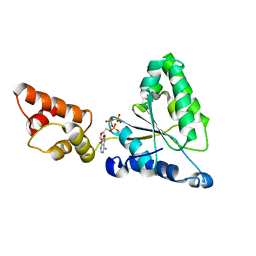 | | Crystal structure of the FtsH ATPase domain with AMP-PNP from Thermus thermophilus | | Descriptor: | ATP-dependent metalloprotease FtsH, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Niwa, H, Tsuchiya, D, Makyio, H, Yoshida, M, Morikawa, K. | | Deposit date: | 2002-07-10 | | Release date: | 2002-11-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Hexameric ring structure of the ATPase domain of the membrane-integrated metalloprotease FtsH from Thermus thermophilus HB8
Structure, 10, 2002
|
|
1IY1
 
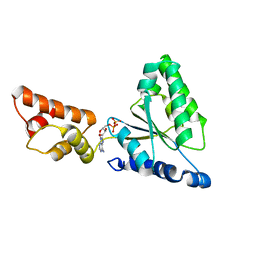 | | Crystal structure of the FtsH ATPase domain with ADP from Thermus thermophilus | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent metalloprotease FtsH | | Authors: | Niwa, H, Tsuchiya, D, Makyio, H, Yoshida, M, Morikawa, K. | | Deposit date: | 2002-07-10 | | Release date: | 2002-11-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Hexameric ring structure of the ATPase domain of the membrane-integrated metalloprotease FtsH from Thermus thermophilus HB8
Structure, 10, 2002
|
|
1IY2
 
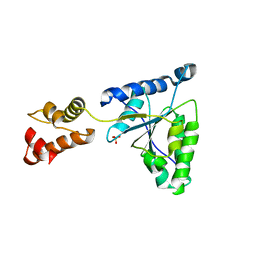 | | Crystal structure of the FtsH ATPase domain from Thermus thermophilus | | Descriptor: | ATP-dependent metalloprotease FtsH, SULFATE ION | | Authors: | Niwa, H, Tsuchiya, D, Makyio, H, Yoshida, M, Morikawa, K. | | Deposit date: | 2002-07-10 | | Release date: | 2002-11-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Hexameric ring structure of the ATPase domain of the membrane-integrated metalloprotease FtsH from Thermus thermophilus HB8
Structure, 10, 2002
|
|
1IY3
 
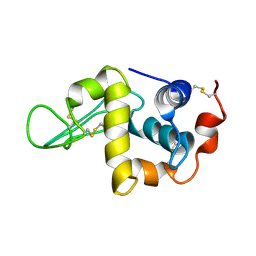 | | Solution Structure of the Human lysozyme at 4 degree C | | Descriptor: | Lysozyme | | Authors: | Kumeta, H, Miura, A, Kobashigawa, Y, Miura, K, Oka, C, Nitta, K, Nemoto, N, Tsuda, S. | | Deposit date: | 2002-07-15 | | Release date: | 2002-07-31 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Low-temperature-induced structural changes in human lysozyme elucidated by three-dimensional NMR spectroscopy
Biochemistry, 42, 2003
|
|
1IY4
 
 | | Solution structure of the human lysozyme at 35 degree C | | Descriptor: | Lysozyme | | Authors: | Kumeta, H, Miura, A, Kobashigawa, Y, Miura, K, Oka, C, Nitta, K, Nemoto, N, Tsuda, S. | | Deposit date: | 2002-07-15 | | Release date: | 2002-07-31 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Low-temperature-induced structural changes in human lysozyme elucidated by three-dimensional NMR spectroscopy
Biochemistry, 42, 2003
|
|
1IY5
 
 | | Solution structure of wild type OMSVP3 | | Descriptor: | OMSVP3 | | Authors: | Hemmi, H, Kumazaki, T, Yamazaki, T, Kojima, S, Yoshida, T, Kyogoku, Y, Katsu, M, Yokosawa, H, Miura, K, Kobayashi, Y. | | Deposit date: | 2002-07-23 | | Release date: | 2003-03-11 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Inhibitory Specificity Change of Ovomucoid Third Domain of the Silver Pheasant upon Introduction of an Engineered Cys14-Cys39 Bond
BIOCHEMISTRY, 42, 2003
|
|
1IY6
 
 | | Solution structure of OMSVP3 variant, P14C/N39C | | Descriptor: | OMSVP3 | | Authors: | Hemmi, H, Kumazaki, T, Yamazaki, T, Kojima, S, Yoshida, T, Kyogoku, Y, Katsu, M, Yokosawa, H, Miura, K, Kobayashi, Y. | | Deposit date: | 2002-07-23 | | Release date: | 2003-03-11 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Inhibitory Specificity Change of Ovomucoid Third Domain of the Silver Pheasant upon Introduction of an Engineered Cys14-Cys39 Bond
BIOCHEMISTRY, 42, 2003
|
|
1IY7
 
 | | Crystal Structure of CPA and sulfamide-based inhibitor complex | | Descriptor: | Carboxypeptidase A, PHENYLALANINE-N-SULFONAMIDE, ZINC ION | | Authors: | Kim, S.J, Woo, J.R, Park, J.D, Kim, D.H, Ryu, S.E. | | Deposit date: | 2002-07-24 | | Release date: | 2003-01-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Sulfamide-Based Inhibitors for Carboxypeptidase A. Novel Type Transition State Analogue Inhibitors for Zinc Proteases
J.Med.Chem., 45, 2002
|
|
1IY8
 
 | | Crystal Structure of Levodione Reductase | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, LEVODIONE REDUCTASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Sogabe, S, Fukami, T, Shiratori, Y, Yoshizumi, A, Wada, M. | | Deposit date: | 2002-07-25 | | Release date: | 2003-05-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Crystal Structure and Stereospecificity of Levodione Reductase from Corynebacterium aquaticum M-13
J.BIOL.CHEM., 278, 2003
|
|
1IY9
 
 | | Crystal structure of spermidine synthase | | Descriptor: | Spermidine synthase | | Authors: | Tan, A.Y, Smith, P.C, Shen, J, Xiao, R, Acton, T, Rost, B, Montelione, G, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2002-07-26 | | Release date: | 2002-10-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of spermidine synthase
To be Published
|
|
1IYB
 
 | |
1IYC
 
 | | Solution structure of antifungal peptide, scarabaecin | | Descriptor: | scarabaecin | | Authors: | Hemmi, H, Ishibashi, J, Tomie, T, Yamakawa, M. | | Deposit date: | 2002-08-05 | | Release date: | 2003-06-24 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for New Pattern of Conserved Amino Acid Residues Related to Chitin-binding in the Antifungal Peptide from the Coconut Rhinoceros Beetle Oryctes rhinoceros
J.BIOL.CHEM., 278, 2003
|
|
