1IHT
 
 | |
1IHU
 
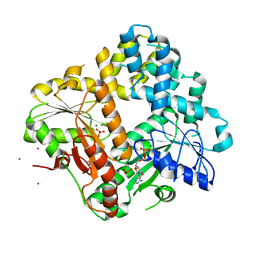 | | CRYSTAL STRUCTURE OF THE ESCHERICHIA COLI ARSENITE-TRANSLOCATING ATPASE IN COMPLEX WITH MG-ADP-ALF3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, ARSENICAL PUMP-DRIVING ATPASE, ... | | Authors: | Zhou, T, Radaev, S, Rosen, B.P, Gatti, D.L. | | Deposit date: | 2001-04-20 | | Release date: | 2001-09-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Conformational changes in four regions of the Escherichia coli ArsA ATPase link ATP hydrolysis to ion translocation.
J.Biol.Chem., 276, 2001
|
|
1IHV
 
 | | SOLUTION STRUCTURE OF THE DNA BINDING DOMAIN OF HIV-1 INTEGRASE, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | HIV-1 INTEGRASE | | Authors: | Clore, G.M, Lodi, P.J, Ernst, J.A, Gronenborn, A.M. | | Deposit date: | 1995-05-12 | | Release date: | 1996-10-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the DNA binding domain of HIV-1 integrase.
Biochemistry, 34, 1995
|
|
1IHW
 
 | | SOLUTION STRUCTURE OF THE DNA BINDING DOMAIN OF HIV-1 INTEGRASE, NMR, 40 STRUCTURES | | Descriptor: | HIV-1 INTEGRASE | | Authors: | Clore, G.M, Lodi, P.J, Ernst, J.A, Gronenborn, A.M. | | Deposit date: | 1995-05-12 | | Release date: | 1996-07-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the DNA binding domain of HIV-1 integrase.
Biochemistry, 34, 1995
|
|
1IHX
 
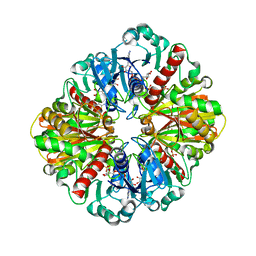 | | Crystal structure of two D-glyceraldehyde-3-phosphate dehydrogenase complexes: a case of asymmetry | | Descriptor: | GLYCERALDEHYDE 3-PHOSPHATE DEHYDROGENASE, SULFATE ION, THIONICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Shen, Y.-Q, Song, S.-Y, Lin, Z.-J. | | Deposit date: | 2001-04-20 | | Release date: | 2002-07-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of D-glyceraldehyde-3-phosphate dehydrogenase complexed with coenzyme analogues.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1IHY
 
 | | GAPDH complexed with ADP-ribose | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, GLYCERALDEHYDE 3-PHOSPHATE DEHYDROGENASE, SULFATE ION | | Authors: | Shen, Y.-Q, Song, S.-Y, Lin, Z.-J. | | Deposit date: | 2001-04-20 | | Release date: | 2002-07-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of D-glyceraldehyde-3-phosphate dehydrogenase complexed with coenzyme analogues.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1IHZ
 
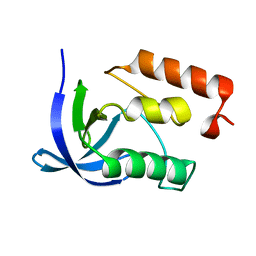 | |
1II0
 
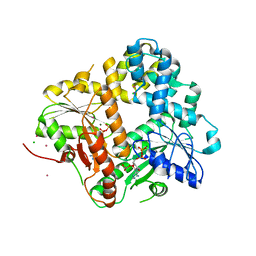 | | CRYSTAL STRUCTURE OF THE ESCHERICHIA COLI ARSENITE-TRANSLOCATING ATPASE | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ARSENICAL PUMP-DRIVING ATPASE, ... | | Authors: | Zhou, T, Radaev, S, Rosen, B.P, Gatti, D.L. | | Deposit date: | 2001-04-20 | | Release date: | 2001-09-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Conformational changes in four regions of the Escherichia coli ArsA ATPase link ATP hydrolysis to ion translocation.
J.Biol.Chem., 276, 2001
|
|
1II1
 
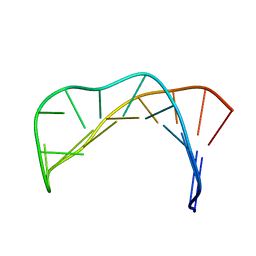 | |
1II2
 
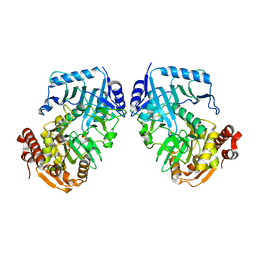 | | Crystal Structure of Phosphoenolpyruvate Carboxykinase (PEPCK) from Trypanosoma cruzi | | Descriptor: | PHOSPHOENOLPYRUVATE CARBOXYKINASE, SULFATE ION | | Authors: | Trapani, S, Linss, J, Goldenberg, S, Fischer, H, Craievich, A.F, Oliva, G. | | Deposit date: | 2001-04-20 | | Release date: | 2001-11-21 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the dimeric phosphoenolpyruvate carboxykinase (PEPCK) from Trypanosoma cruzi at 2 A resolution.
J.Mol.Biol., 313, 2001
|
|
1II3
 
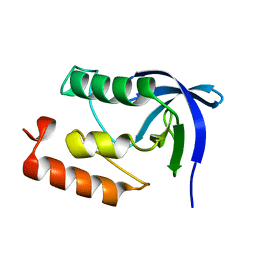 | |
1II4
 
 | | CRYSTAL STRUCTURE OF SER252TRP APERT MUTANT FGF RECEPTOR 2 (FGFR2) IN COMPLEX WITH FGF2 | | Descriptor: | FIBROBLAST GROWTH FACTOR RECEPTOR 2, HEPARIN-BINDING GROWTH FACTOR 2 | | Authors: | Ibrahimi, O.A, Eliseenkova, A.V, Plotnikov, A.N, Ornitz, D.M, Mohammadi, M. | | Deposit date: | 2001-04-20 | | Release date: | 2001-05-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for fibroblast growth factor receptor 2 activation in Apert syndrome.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1II5
 
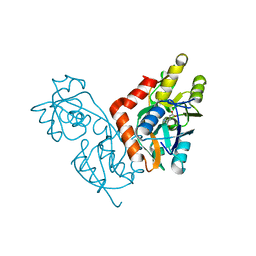 | |
1II6
 
 | | Crystal Structure of the Mitotic Kinesin Eg5 in Complex with Mg-ADP. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, KINESIN-RELATED MOTOR PROTEIN Eg5, MAGNESIUM ION, ... | | Authors: | Turner, J, Anderson, R, Guo, J, Beraud, C, Sakowicz, R, Fletterick, R. | | Deposit date: | 2001-04-20 | | Release date: | 2001-07-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the mitotic spindle kinesin Eg5 reveals a novel conformation of the neck-linker.
J.Biol.Chem., 276, 2001
|
|
1II7
 
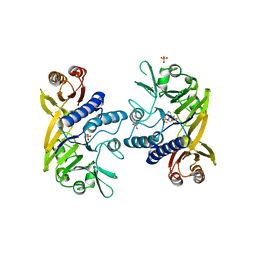 | | Crystal structure of P. furiosus Mre11 with manganese and dAMP | | Descriptor: | 2'-DEOXYADENOSINE-5'-MONOPHOSPHATE, MANGANESE (II) ION, Mre11 nuclease, ... | | Authors: | Hopfner, K.-P, Karcher, A, Craig, L, Woo, T.T, Carney, J.P, Tainer, J.A. | | Deposit date: | 2001-04-20 | | Release date: | 2001-05-30 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural biochemistry and interaction architecture of the DNA double-strand break repair Mre11 nuclease and Rad50-ATPase.
Cell(Cambridge,Mass.), 105, 2001
|
|
1II8
 
 | | Crystal structure of the P. furiosus Rad50 ATPase domain | | Descriptor: | PHOSPHATE ION, Rad50 ABC-ATPase | | Authors: | Hopfner, K.-P, Karcher, A, Craig, L, Woo, T.T, Carney, J.P, Tainer, J.A. | | Deposit date: | 2001-04-20 | | Release date: | 2001-05-30 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Structural biochemistry and interaction architecture of the DNA double-strand break repair Mre11 nuclease and Rad50-ATPase.
Cell(Cambridge,Mass.), 105, 2001
|
|
1II9
 
 | | CRYSTAL STRUCTURE OF THE ESCHERICHIA COLI ARSENITE-TRANSLOCATING ATPASE IN COMPLEX WITH AMP-PNP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ARSENICAL PUMP-DRIVING ATPASE, CADMIUM ION, ... | | Authors: | Zhou, T, Radaev, S, Gatti, D.L, Rosen, B.P. | | Deposit date: | 2001-04-21 | | Release date: | 2001-09-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Conformational changes in four regions of the Escherichia coli ArsA ATPase link ATP hydrolysis to ion translocation.
J.Biol.Chem., 276, 2001
|
|
1IIB
 
 | | CRYSTAL STRUCTURE OF IIBCELLOBIOSE FROM ESCHERICHIA COLI | | Descriptor: | ENZYME IIB OF THE CELLOBIOSE-SPECIFIC PHOSPHOTRANSFERASE SYSTEM | | Authors: | Van Montfort, R.L.M, Pijning, T, Kalk, K.H, Reizer, J, Saier, M.H, Thunnissen, M.M.G.M, Robillard, G.T, Dijkstra, B.W. | | Deposit date: | 1996-12-23 | | Release date: | 1997-12-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure of an energy-coupling protein from bacteria, IIBcellobiose, reveals similarity to eukaryotic protein tyrosine phosphatases.
Structure, 5, 1997
|
|
1IIC
 
 | |
1IID
 
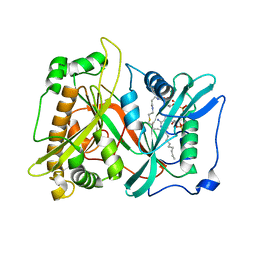 | | Crystal Structure of Saccharomyces cerevisiae N-myristoyltransferase with Bound S-(2-oxo)pentadecylCoA and the Octapeptide GLYASKLA | | Descriptor: | NICKEL (II) ION, Octapeptide GLYASKLA, Peptide N-myristoyltransferase, ... | | Authors: | Farazi, T.A, Gordon, J.I, Waksman, G. | | Deposit date: | 2001-04-22 | | Release date: | 2001-05-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of Saccharomyces cerevisiae N-myristoyltransferase with bound myristoylCoA and peptide provide insights about substrate recognition and catalysis.
Biochemistry, 40, 2001
|
|
1IIE
 
 | |
1IIG
 
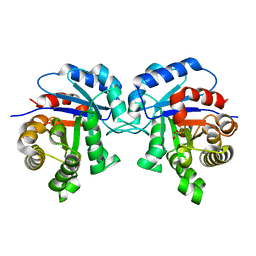 | | STRUCTURE OF TRYPANOSOMA BRUCEI BRUCEI TRIOSEPHOSPHATE ISOMERASE COMPLEXED WITH 3-PHOSPHONOPROPIONATE | | Descriptor: | 3-PHOSPHONOPROPANOIC ACID, TRIOSEPHOSPHATE ISOMERASE | | Authors: | Noble, M.E, Wierenga, R.K, Lambeir, A.M, Opperdoes, F.R, Thunnissen, A.M, Kalk, K.H, Groendijk, H, Hol, W.G.J. | | Deposit date: | 2001-04-23 | | Release date: | 2001-05-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The adaptability of the active site of trypanosomal triosephosphate isomerase as observed in the crystal structures of three different complexes.
Proteins, 10, 1991
|
|
1IIH
 
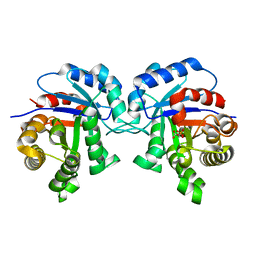 | | STRUCTURE OF TRYPANOSOMA BRUCEI BRUCEI TRIOSEPHOSPHATE ISOMERASE COMPLEXED WITH 3-PHOSPHOGLYCERATE | | Descriptor: | 3-PHOSPHOGLYCERIC ACID, TRIOSEPHOSPHATE ISOMERASE | | Authors: | Noble, M.E, Wierenga, R.K, Lambeir, A.M, Opperdoes, F.R, Thunnissen, A.M, Kalk, K.H, Groendijk, H, Hol, W.G.J. | | Deposit date: | 2001-04-23 | | Release date: | 2001-05-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The adaptability of the active site of trypanosomal triosephosphate isomerase as observed in the crystal structures of three different complexes.
Proteins, 10, 1991
|
|
1III
 
 | | CRYSTAL STRUCTURE OF THE TRANSTHYRETIN MUTANT TTR Y114C-DATA COLLECTED AT ROOM TEMPERATURE | | Descriptor: | BETA-MERCAPTOETHANOL, TRANSTHYRETIN | | Authors: | Eneqvist, T, Olofsson, A, Ando, Y, Lundgren, E, Sauer-Eriksson, A.E. | | Deposit date: | 2001-04-23 | | Release date: | 2002-11-15 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Disulfide-Bond Formation in the Transthyretin Mutant Y114C Prevents Amyloid Fibril Formation in Vivo and in Vitro
Biochemistry, 41, 2002
|
|
1IIJ
 
 | |
