1IFI
 
 | |
1IFJ
 
 | |
1IFK
 
 | |
1IFL
 
 | |
1IFM
 
 | |
1IFN
 
 | |
1IFP
 
 | |
1IFQ
 
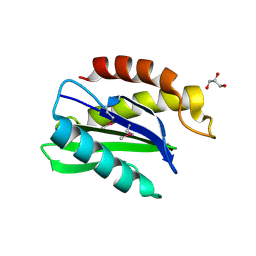 | | Sec22b N-terminal domain | | Descriptor: | GLYCEROL, vesicle trafficking protein Sec22b | | Authors: | Gonzalez Jr, L.C, Weis, W.I, Scheller, R.H. | | Deposit date: | 2001-04-13 | | Release date: | 2001-05-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A novel snare N-terminal domain revealed by the crystal structure of Sec22b.
J.Biol.Chem., 276, 2001
|
|
1IFR
 
 | |
1IFS
 
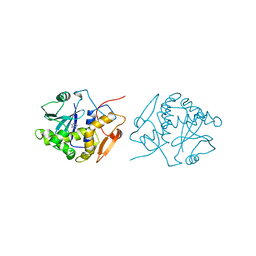 | | RICIN A-CHAIN (RECOMBINANT) COMPLEX WITH ADENOSINE (ADENOSINE BECOMES ADENINE IN THE COMPLEX) | | Descriptor: | ADENINE, RICIN | | Authors: | Weston, S.A, Tucker, A.D, Thatcher, D.R, Derbyshire, D.J, Pauptit, R.A. | | Deposit date: | 1996-07-05 | | Release date: | 1998-01-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structure of recombinant ricin A-chain at 1.8 A resolution.
J.Mol.Biol., 244, 1994
|
|
1IFT
 
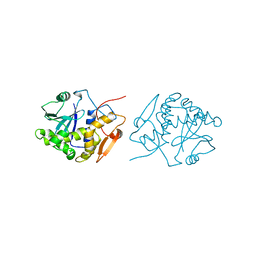 | | RICIN A-CHAIN (RECOMBINANT) | | Descriptor: | RICIN | | Authors: | Weston, S.A, Tucker, A.D, Thatcher, D.R, Derbyshire, D.J, Pauptit, R.A. | | Deposit date: | 1996-07-05 | | Release date: | 1998-01-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray structure of recombinant ricin A-chain at 1.8 A resolution.
J.Mol.Biol., 244, 1994
|
|
1IFU
 
 | | RICIN A-CHAIN (RECOMBINANT) COMPLEX WITH FORMYCIN | | Descriptor: | (1S)-1-(7-amino-1H-pyrazolo[4,3-d]pyrimidin-3-yl)-1,4-anhydro-D-ribitol, RICIN | | Authors: | Weston, S.A, Tucker, A.D, Thatcher, D.R, Derbyshire, D.J, Pauptit, R.A. | | Deposit date: | 1996-07-05 | | Release date: | 1998-01-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-ray structure of recombinant ricin A-chain at 1.8 A resolution.
J.Mol.Biol., 244, 1994
|
|
1IFV
 
 | |
1IFW
 
 | | SOLUTION STRUCTURE OF C-TERMINAL DOMAIN OF POLY(A) BINDING PROTEIN FROM SACCHAROMYCES CEREVISIAE | | Descriptor: | POLYADENYLATE-BINDING PROTEIN, CYTOPLASMIC AND NUCLEAR | | Authors: | Kozlov, G, Siddiqui, N, Coillet-Matillon, S, Sprules, T, Ekiel, I, Gehring, K. | | Deposit date: | 2001-04-13 | | Release date: | 2002-07-24 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the orphan PABC domain from Saccharomyces cerevisiae poly(A)-binding protein.
J.Biol.Chem., 277, 2002
|
|
1IFX
 
 | | CRYSTAL STRUCTURE OF NH3-DEPENDENT NAD+ SYNTHETASE FROM BACILLUS SUBTILIS COMPLEXED WITH TWO MOLECULES DEAMIDO-NAD | | Descriptor: | NH(3)-DEPENDENT NAD(+) SYNTHETASE, NICOTINIC ACID ADENINE DINUCLEOTIDE | | Authors: | Devedjiev, Y, Symersky, J, Singh, R, Brouillette, W, Muccio, D, Jedrzejas, M, Brouillette, C, DeLucas, L. | | Deposit date: | 2001-04-13 | | Release date: | 2001-06-06 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Stabilization of active-site loops in NH3-dependent NAD+ synthetase from Bacillus subtilis.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1IFY
 
 | |
1IG0
 
 | | Crystal Structure of yeast Thiamin Pyrophosphokinase | | Descriptor: | 3-(4-AMINO-2-METHYL-PYRIMIDIN-5-YLMETHYL)-5-(2-HYDROXY-ETHYL)-4-METHYL-THIAZOL-3-IUM, Thiamin pyrophosphokinase | | Authors: | Baker, L.-J, Dorocke, J.A, Harris, R.A, Timm, D.E. | | Deposit date: | 2001-04-16 | | Release date: | 2001-06-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of yeast thiamin pyrophosphokinase.
Structure, 9, 2001
|
|
1IG1
 
 | | 1.8A X-Ray structure of ternary complex of a catalytic domain of death-associated protein kinase with ATP analogue and Mn. | | Descriptor: | MANGANESE (II) ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, death-associated protein kinase | | Authors: | Tereshko, V, Teplova, M, Brunzelle, J, Watterson, D.M, Egli, M. | | Deposit date: | 2001-04-16 | | Release date: | 2002-04-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of the catalytic domain of human protein kinase associated with apoptosis and tumor suppression.
Nat.Struct.Biol., 8, 2001
|
|
1IG3
 
 | | Mouse Thiamin Pyrophosphokinase Complexed with Thiamin | | Descriptor: | 3-(4-AMINO-2-METHYL-PYRIMIDIN-5-YLMETHYL)-5-(2-HYDROXY-ETHYL)-4-METHYL-THIAZOL-3-IUM, SULFATE ION, thiamin pyrophosphokinase | | Authors: | Timm, D.E, Liu, J, Baker, L.-J, Harris, R.A. | | Deposit date: | 2001-04-16 | | Release date: | 2001-04-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of thiamin pyrophosphokinase.
J.Mol.Biol., 310, 2001
|
|
1IG4
 
 | | Solution Structure of the Methyl-CpG-Binding Domain of Human MBD1 in Complex with Methylated DNA | | Descriptor: | 5'-D(*GP*TP*AP*TP*CP*(5CM)P*GP*GP*AP*TP*AP*C)-3', Methyl-CpG Binding Protein | | Authors: | Ohki, I, Shimotake, N, Fujita, N, Jee, J.-G, Ikegami, T, Nakao, M, Shirakawa, M. | | Deposit date: | 2001-04-17 | | Release date: | 2001-05-30 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the methyl-CpG binding domain of human MBD1 in complex with methylated DNA.
Cell(Cambridge,Mass.), 105, 2001
|
|
1IG5
 
 | | BOVINE CALBINDIN D9K BINDING MG2+ | | Descriptor: | MAGNESIUM ION, VITAMIN D-DEPENDENT CALCIUM-BINDING PROTEIN, INTESTINAL | | Authors: | Andersson, E.M, Svensson, L.A. | | Deposit date: | 2001-04-17 | | Release date: | 2001-04-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis for the negative allostery between Ca(2+)- and Mg(2+)-binding in the intracellular Ca(2+)-receptor calbindin D9k.
Protein Sci., 6, 1997
|
|
1IG6
 
 | | HUMAN MRF-2 DOMAIN, NMR, 11 STRUCTURES | | Descriptor: | MODULATOR RECOGNITION FACTOR 2 | | Authors: | Lin, D, Tsui, V, Case, D, Yuan, Y.C, Chen, Y. | | Deposit date: | 2001-04-17 | | Release date: | 2001-04-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | HUMAN MRF-2 DOMAIN, NMR, 11 STRUCTURES
To be Published
|
|
1IG7
 
 | | Msx-1 Homeodomain/DNA Complex Structure | | Descriptor: | 5'-D(*CP*AP*CP*TP*AP*AP*TP*TP*GP*AP*AP*GP*G)-3', 5'-D(P*TP*CP*CP*TP*TP*CP*AP*AP*TP*TP*AP*GP*TP*GP*AP*C)-3', Homeotic protein Msx-1 | | Authors: | Hovde, S, Abate-Shen, C, Geiger, J.H. | | Deposit date: | 2001-04-17 | | Release date: | 2001-04-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the Msx-1 homeodomain/DNA complex
Biochemistry, 40, 2001
|
|
1IG8
 
 | | Crystal Structure of Yeast Hexokinase PII with the correct amino acid sequence | | Descriptor: | SULFATE ION, hexokinase PII | | Authors: | Kuser, P.R, Krauchenco, S, Antunes, O.A, Polikarpov, I. | | Deposit date: | 2001-04-17 | | Release date: | 2001-05-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The high resolution crystal structure of yeast hexokinase PII with the correct primary sequence provides new insights into its mechanism of action.
J.Biol.Chem., 275, 2000
|
|
1IG9
 
 | | Structure of the Replicating Complex of a Pol Alpha Family DNA Polymerase | | Descriptor: | 5'-D(*AP*CP*AP*GP*GP*TP*AP*AP*GP*CP*AP*GP*TP*CP*CP*GP*CP*G)-3', 5'-D(*GP*CP*GP*GP*AP*CP*TP*GP*CP*TP*TP*AP*CP*(DOC))-3', CALCIUM ION, ... | | Authors: | Franklin, M.C, Wang, J, Steitz, T.A. | | Deposit date: | 2001-04-17 | | Release date: | 2001-06-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the Replicating Complex of a Pol Alpha Family DNA Polymerase
Cell(Cambridge,Mass.), 105, 2001
|
|
