1IB2
 
 | |
1IB4
 
 | |
1IB5
 
 | | X-RAY 3D STRUCTURE OF P.LEIOGNATHI CU,ZN SOD MUTANT W83Y | | Descriptor: | COPPER (II) ION, CU,ZN SUPEROXIDE DISMUTASE, ZINC ION | | Authors: | Stroppolo, M.E, Pesce, A, D'Orazio, M, O'Neill, P, Bordo, D, Rosano, C, Milani, M, Battistoni, A, Bolognesi, M, Desideri, A. | | Deposit date: | 2001-03-27 | | Release date: | 2001-05-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Single mutations at the subunit interface modulate copper reactivity in Photobacterium leiognathi Cu,Zn superoxide dismutase.
J.Mol.Biol., 308, 2001
|
|
1IB6
 
 | | CRYSTAL STRUCTURE OF R153C E. COLI MALATE DEHYDROGENASE | | Descriptor: | MALATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION | | Authors: | Bell, J.K, Yennawar, H.P, Wright, S.K, Thompson, J.R, Viola, R.E, Banaszak, L.J. | | Deposit date: | 2001-03-27 | | Release date: | 2001-09-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Analyses of a Malate Dehydrogenase with a Variable Active Site
J.Biol.Chem., 276, 2001
|
|
1IB7
 
 | | SOLUTION STRUCTURE OF F35Y MUTANT OF RAT FERRO CYTOCHROME B5, A CONFORMATION, ENSEMBLE OF 20 STRUCTURES | | Descriptor: | CYTOCHROME B5, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Shahzad, N, Dangi, B, Blankman, J.I, Guiles, R.D. | | Deposit date: | 2001-03-27 | | Release date: | 2001-04-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | MUTAGENIC MODULATION OF THE ENTROPY CHANGE ON OXIDATION OF CYTOCHROME B5: AN ANALYSIS OF THE CONTRIBUTION OF CONFORMATIONAL ENTROPY
To be Published
|
|
1IB8
 
 | | SOLUTION STRUCTURE AND FUNCTION OF A CONSERVED PROTEIN SP14.3 ENCODED BY AN ESSENTIAL STREPTOCOCCUS PNEUMONIAE GENE | | Descriptor: | CONSERVED PROTEIN SP14.3 | | Authors: | Yu, L, Gunasekera, A.H, Mack, J, Olejniczak, E.T, Chovan, L.E, Ruan, X, Towne, D.L, Lerner, C.G, Fesik, S.W. | | Deposit date: | 2001-03-27 | | Release date: | 2002-03-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | SOLUTION STRUCTURE AND FUNCTION OF A CONSERVED PROTEIN SP14.3 ENCODED BY AN ESSENTIAL STREPTOCOCCUS PNEUMONIAE GENE
J.Mol.Biol., 311, 2001
|
|
1IB9
 
 | |
1IBA
 
 | | GLUCOSE PERMEASE (DOMAIN IIB), NMR, 11 STRUCTURES | | Descriptor: | GLUCOSE PERMEASE | | Authors: | Eberstadt, M, Grdadolnik, S.G, Gemmecker, G, Kessler, H, Buhr, A, Erni, B. | | Deposit date: | 1996-03-23 | | Release date: | 1996-10-14 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the IIB domain of the glucose transporter of Escherichia coli.
Biochemistry, 35, 1996
|
|
1IBB
 
 | | X-RAY 3D STRUCTURE OF P.LEIOGNATHI CU,ZN SOD MUTANT W83F | | Descriptor: | COPPER (II) ION, CU,ZN SUPEROXIDE DISMUTASE, ZINC ION | | Authors: | Stroppolo, M.E, Pesce, A, D'Orazio, M, O'Neill, P, Bordo, D, Rosano, C, Milani, M, Battistoni, A, Bolognesi, M, Desideri, A. | | Deposit date: | 2001-03-28 | | Release date: | 2001-05-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Single mutations at the subunit interface modulate copper reactivity in Photobacterium leiognathi Cu,Zn superoxide dismutase.
J.Mol.Biol., 308, 2001
|
|
1IBC
 
 | |
1IBD
 
 | | X-RAY 3D STRUCTURE OF P.LEIOGNATHI CU,ZN SOD MUTANT V29A | | Descriptor: | COPPER (II) ION, CU,ZN SUPEROXIDE DISMUTASE, ZINC ION | | Authors: | Stroppolo, M.E, Pesce, A, D'Orazio, M, O'Neill, P, Bordo, D, Rosano, C, Milani, M, Battistoni, A, Bolognesi, M, Desideri, A. | | Deposit date: | 2001-03-28 | | Release date: | 2001-05-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Single mutations at the subunit interface modulate copper reactivity in Photobacterium leiognathi Cu,Zn superoxide dismutase.
J.Mol.Biol., 308, 2001
|
|
1IBE
 
 | |
1IBF
 
 | | X-RAY 3D STRUCTURE OF P.LEIOGNATHI CU,ZN SOD MUTANT V29G | | Descriptor: | COPPER (II) ION, CU,ZN SUPEROXIDE DISMUTASE, ZINC ION | | Authors: | Stroppolo, M.E, Pesce, A, D'Orazio, M, O'Neill, P, Bordo, D, Rosano, C, Milani, M, Battistoni, A, Bolognesi, M, Desideri, A. | | Deposit date: | 2001-03-28 | | Release date: | 2001-05-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Single mutations at the subunit interface modulate copper reactivity in Photobacterium leiognathi Cu,Zn superoxide dismutase.
J.Mol.Biol., 308, 2001
|
|
1IBG
 
 | |
1IBH
 
 | | X-RAY 3D STRUCTURE OF P.LEIOGNATHI CU,ZN SOD MUTANT M41I | | Descriptor: | COPPER (II) ION, CU,ZN SUPEROXIDE DISMUTASE, ZINC ION | | Authors: | Stroppolo, M.E, Pesce, A, D'Orazio, M, O'Neill, P, Bordo, D, Rosano, C, Milani, M, Battistoni, A, Bolognesi, M, Desideri, A. | | Deposit date: | 2001-03-28 | | Release date: | 2001-05-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Single mutations at the subunit interface modulate copper reactivity in Photobacterium leiognathi Cu,Zn superoxide dismutase.
J.Mol.Biol., 308, 2001
|
|
1IBI
 
 | | QUAIL CYSTEINE AND GLYCINE-RICH PROTEIN, NMR, 15 MINIMIZED MODEL STRUCTURES | | Descriptor: | CYSTEINE-RICH PROTEIN 2, ZINC ION | | Authors: | Schuler, W, Kloiber, K, Matt, T, Bister, K, Konrat, R. | | Deposit date: | 2001-03-28 | | Release date: | 2001-09-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Application of cross-correlated NMR spin relaxation to the zinc-finger protein CRP2(LIM2): evidence for collective motions in LIM domains.
Biochemistry, 40, 2001
|
|
1IBJ
 
 | | Crystal structure of cystathionine beta-lyase from Arabidopsis thaliana | | Descriptor: | CARBONATE ION, CYSTATHIONINE BETA-LYASE, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Breitinger, U, Clausen, T, Messerschmidt, A. | | Deposit date: | 2001-03-28 | | Release date: | 2001-04-04 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The three-dimensional structure of cystathionine beta-lyase from Arabidopsis and its substrate specificity
Plant Physiol., 126, 2001
|
|
1IBK
 
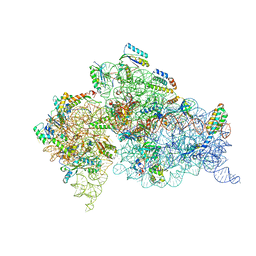 | | STRUCTURE OF THE THERMUS THERMOPHILUS 30S RIBOSOMAL SUBUNIT IN COMPLEX WITH THE ANTIBIOTIC PAROMOMYCIN | | Descriptor: | 16S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Ogle, J.M, Brodersen, D.E, Clemons Jr, W.M, Tarry, M.J, Carter, A.P, Ramakrishnan, V. | | Deposit date: | 2001-03-28 | | Release date: | 2001-05-04 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | Recognition of cognate transfer RNA by the 30S ribosomal subunit.
Science, 292, 2001
|
|
1IBL
 
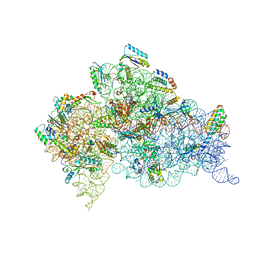 | | STRUCTURE OF THE THERMUS THERMOPHILUS 30S RIBOSOMAL SUBUNIT IN COMPLEX WITH A MESSENGER RNA FRAGMENT AND COGNATE TRANSFER RNA ANTICODON STEM-LOOP BOUND AT THE A SITE AND WITH THE ANTIBIOTIC PAROMOMYCIN | | Descriptor: | 16S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Ogle, J.M, Brodersen, D.E, Clemons Jr, W.M, Tarry, M.J, Carter, A.P, Ramakrishnan, V. | | Deposit date: | 2001-03-28 | | Release date: | 2001-05-04 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Recognition of cognate transfer RNA by the 30S ribosomal subunit.
Science, 292, 2001
|
|
1IBM
 
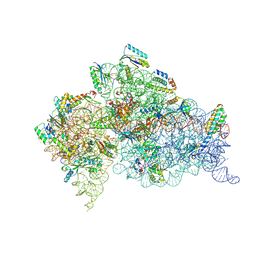 | | STRUCTURE OF THE THERMUS THERMOPHILUS 30S RIBOSOMAL SUBUNIT IN COMPLEX WITH A MESSENGER RNA FRAGMENT AND COGNATE TRANSFER RNA ANTICODON STEM-LOOP BOUND AT THE A SITE | | Descriptor: | 16S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Ogle, J.M, Brodersen, D.E, Clemons Jr, W.M, Tarry, M.J, Carter, A.P, Ramakrishnan, V. | | Deposit date: | 2001-03-28 | | Release date: | 2001-05-04 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | Recognition of cognate transfer RNA by the 30S ribosomal subunit.
Science, 292, 2001
|
|
1IBN
 
 | |
1IBO
 
 | |
1IBQ
 
 | | ASPERGILLOPEPSIN FROM ASPERGILLUS PHOENICIS | | Descriptor: | ASPERGILLOPEPSIN, ZINC ION, alpha-D-mannopyranose | | Authors: | Cho, S.W, Shin, W. | | Deposit date: | 2001-03-28 | | Release date: | 2001-07-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Structure of aspergillopepsin I from Aspergillus phoenicis: variations of the S1'-S2 subsite in aspartic proteinases.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1IBR
 
 | | COMPLEX OF RAN WITH IMPORTIN BETA | | Descriptor: | GTP-binding nuclear protein RAN, Importin beta-1 subunit, MAGNESIUM ION, ... | | Authors: | Vetter, I.R, Arndt, A, Kutay, U, Goerlich, D, Wittinghofer, A. | | Deposit date: | 1999-05-14 | | Release date: | 1999-06-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural view of the Ran-Importin beta interaction at 2.3 A resolution
Cell(Cambridge,Mass.), 97, 1999
|
|
1IBS
 
 | | PHOSPHORIBOSYLDIPHOSPHATE SYNTHETASE IN COMPLEX WITH CADMIUM IONS | | Descriptor: | CADMIUM ION, METHYL PHOSPHONIC ACID ADENOSINE ESTER, RIBOSE-PHOSPHATE PYROPHOSPHOKINASE, ... | | Authors: | Eriksen, T.A, Kadziola, A, Larsen, S. | | Deposit date: | 2001-03-29 | | Release date: | 2002-02-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Binding of cations in Bacillus subtilis phosphoribosyldiphosphate synthetase and their role in catalysis.
Protein Sci., 11, 2002
|
|
