6TY9
 
 | | In situ structure of BmCPV RNA dependent RNA polymerase at initiation state | | 分子名称: | MAGNESIUM ION, Non-template RNA (5'-D(*(GTA))-R(P*GP*UP*AP*AP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*U)-3'), RNA-dependent RNA Polymerase, ... | | 著者 | Cui, Y.X, Zhang, Y.N, Sun, J.C, Zhou, Z.H. | | 登録日 | 2019-08-08 | | 公開日 | 2019-11-20 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Conservative transcription in three steps visualized in a double-stranded RNA virus.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6TZ1
 
 | | In situ structure of BmCPV RNA-dependent RNA polymerase at early-elongation state | | 分子名称: | Non-template RNA (5'-D(*(GTA))-R(P*GP*UP*A)-3'), RNA-dependent RNA Polymerase, Template RNA (5'-R(P*AP*GP*AP*AP*AP*AP*AP*AP*AP*AP*A)-3'), ... | | 著者 | Cui, Y.X, Zhang, Y.N, Sun, J.C, Zhou, Z.H. | | 登録日 | 2019-08-09 | | 公開日 | 2019-11-20 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Conservative transcription in three steps visualized in a double-stranded RNA virus.
Nat.Struct.Mol.Biol., 26, 2019
|
|
3H4J
 
 | | crystal structure of pombe AMPK KDAID fragment | | 分子名称: | SNF1-like protein kinase ssp2 | | 著者 | Chen, L, Jiao, Z.-H, Zheng, L.-S, Zhang, Y.-Y, Xie, S.-T, Wang, Z.-X, Wu, J.-W. | | 登録日 | 2009-04-20 | | 公開日 | 2009-06-02 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structural insight into the autoinhibition mechanism of AMP-activated protein kinase
Nature, 459, 2009
|
|
6TY8
 
 | | In situ structure of BmCPV RNA dependent RNA polymerase at quiescent state | | 分子名称: | P1-7-METHYLGUANOSINE-P3-ADENOSINE-5',5'-TRIPHOSPHATE, RNA-dependent RNA Polymerase, Viral structural protein 4 | | 著者 | Cui, Y.X, Zhang, Y.N, Sun, J.C, Zhou, Z.H. | | 登録日 | 2019-08-08 | | 公開日 | 2019-11-20 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Conservative transcription in three steps visualized in a double-stranded RNA virus.
Nat.Struct.Mol.Biol., 26, 2019
|
|
5X58
 
 | | Prefusion structure of SARS-CoV spike glycoprotein, conformation 1 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Yuan, Y, Cao, D, Zhang, Y, Ma, J, Qi, J, Wang, Q, Lu, G, Wu, Y, Yan, J, Shi, Y, Zhang, X, Gao, G.F. | | 登録日 | 2017-02-15 | | 公開日 | 2017-05-03 | | 最終更新日 | 2020-07-29 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains
Nat Commun, 8, 2017
|
|
1SQ5
 
 | | Crystal Structure of E. coli Pantothenate kinase | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, PANTOTHENOIC ACID, Pantothenate kinase | | 著者 | Ivey, R.A, Zhang, Y.-M, Virga, K.G, Hevener, K, Lee, R.E, Rock, C.O, Jackowski, S, Park, H.-W. | | 登録日 | 2004-03-17 | | 公開日 | 2004-09-28 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | The structure of the pantothenate kinase.ADP.pantothenate ternary complex reveals the relationship between the binding sites for substrate, allosteric regulator, and antimetabolites.
J.Biol.Chem., 279, 2004
|
|
8XMD
 
 | |
5X5B
 
 | | Prefusion structure of SARS-CoV spike glycoprotein, conformation 2 | | 分子名称: | Spike glycoprotein | | 著者 | Yuan, Y, Cao, D, Zhang, Y, Ma, J, Qi, J, Wang, Q, Lu, G, Wu, Y, Yan, J, Shi, Y, Zhang, X, Gao, G.F. | | 登録日 | 2017-02-15 | | 公開日 | 2017-05-03 | | 最終更新日 | 2017-05-24 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains
Nat Commun, 8, 2017
|
|
3L5R
 
 | |
8XME
 
 | |
3L5U
 
 | |
3VFE
 
 | | Virtual Screening and X-Ray Crystallography for Human Kallikrein 6 Inhibitors with an Amidinothiophene P1 Group | | 分子名称: | 4-{[(3R)-3-{[(7-methoxynaphthalen-2-yl)sulfonyl](thiophen-3-ylmethyl)amino}-2-oxopyrrolidin-1-yl]methyl}thiophene-2-carboximidamide, Kallikrein-6 | | 著者 | Chen, X, Zhang, Y, Xia, T, Wang, R. | | 登録日 | 2012-01-09 | | 公開日 | 2012-11-21 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.88 Å) | | 主引用文献 | Virtual Screening and X-ray Crystallography for Human Kallikrein 6 Inhibitors with an Amidinothiophene P1 Group.
Acs Med.Chem.Lett., 3, 2012
|
|
3L5S
 
 | |
5X5C
 
 | | Prefusion structure of MERS-CoV spike glycoprotein, conformation 1 | | 分子名称: | S protein | | 著者 | Yuan, Y, Cao, D, Zhang, Y, Ma, J, Qi, J, Wang, Q, Lu, G, Wu, Y, Yan, J, Shi, Y, Zhang, X, Gao, G.F. | | 登録日 | 2017-02-15 | | 公開日 | 2017-05-03 | | 最終更新日 | 2017-05-24 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains
Nat Commun, 8, 2017
|
|
3L5V
 
 | |
3L5P
 
 | |
6LIG
 
 | |
6BM0
 
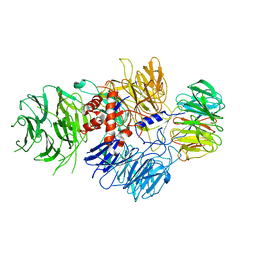 | | Cryo-EM structure of human CPSF-160-WDR33 complex at 3.8 A resolution | | 分子名称: | Cleavage and polyadenylation specificity factor subunit 1, pre-mRNA 3' end processing protein WDR33 | | 著者 | Sun, Y, Zhang, Y, Hamilton, K, Walz, T, Tong, L. | | 登録日 | 2017-11-12 | | 公開日 | 2017-11-22 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Molecular basis for the recognition of the human AAUAAA polyadenylation signal.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
1C1J
 
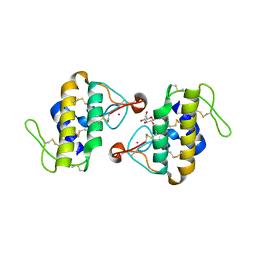 | | STRUCTURE OF CADMIUM-SUBSTITUTED PHOSPHOLIPASE A2 FROM AGKISTRONDON HALYS PALLAS AT 2.8 ANGSTROMS RESOLUTION | | 分子名称: | BASIC PHOSPHOLIPASE A2, CADMIUM ION, octyl beta-D-glucopyranoside | | 著者 | Zhang, H.-l, Zhang, Y.-q, Song, S.-y, Zhou, Y, Lin, Z.-j. | | 登録日 | 1999-07-22 | | 公開日 | 2002-07-05 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structure of Cadmium-substituted Phospholipase A2 from Agkistrodon halys
Pallas at 2.8 Angstroms Resolution
Protein Pept.Lett., 6, 1999
|
|
4IN0
 
 | |
5ZIU
 
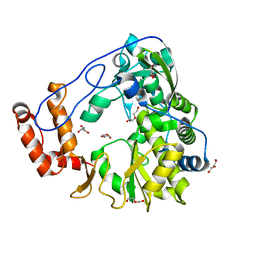 | | Crystal structure of human Entervirus D68 RdRp | | 分子名称: | DI(HYDROXYETHYL)ETHER, GLYCEROL, RdRp | | 著者 | Wang, M.L, Zhang, Y, Chen, Y.P, Lu, D.R, Jiang, H, Chen, Y.J, Li, L, Zhang, C.H, Shi, Q.L, Su, D. | | 登録日 | 2018-03-17 | | 公開日 | 2019-04-17 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.147 Å) | | 主引用文献 | Crystal structure of human Entervirus D68 RdRp in complex with NADPH
To Be Published
|
|
1RQJ
 
 | | Active Conformation of Farnesyl Pyrophosphate Synthase Bound to Isopentyl Pyrophosphate and Risedronate | | 分子名称: | 1-HYDROXY-2-(3-PYRIDINYL)ETHYLIDENE BIS-PHOSPHONIC ACID, 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, Geranyltranstransferase, ... | | 著者 | Hosfield, D.J, Zhang, Y, Dougan, D.R, Brooun, A, Tari, L.W, Swanson, R.V, Finn, J. | | 登録日 | 2003-12-05 | | 公開日 | 2004-03-02 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Structural basis for bisphosphonate-mediated inhibition of isoprenoid biosynthesis
J.Biol.Chem., 279, 2004
|
|
1UO5
 
 | | Structure Based Engineering of Internal Molecular Surfaces Of Four Helix Bundles | | 分子名称: | CHLORIDE ION, GENERAL CONTROL PROTEIN GCN4, iodobenzene | | 著者 | Yadav, M.K, Redman, J.E, Alvarez-Gutierrez, J.M, Zhang, Y. | | 登録日 | 2003-09-15 | | 公開日 | 2004-10-13 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.07 Å) | | 主引用文献 | Structure-Based Engineering of Internal Cavities in Coiled-Coil Peptides
Biochemistry, 44, 2005
|
|
1UNU
 
 | | Structure Based Engineering of Internal Molecular Surfaces Of Four Helix Bundles | | 分子名称: | CHLORIDE ION, GENERAL CONTROL PROTEIN GCN4 | | 著者 | Yadav, M.K, Redman, J.E, Alvarez-Gutierrez, J.M, Zhang, Y, Stout, C.D, Ghadiri, M.R. | | 登録日 | 2003-09-15 | | 公開日 | 2004-10-13 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.07 Å) | | 主引用文献 | Structure-Based Engineering of Internal Cavities in Coiled-Coil Peptides
Biochemistry, 44, 2005
|
|
5X5F
 
 | | Prefusion structure of MERS-CoV spike glycoprotein, conformation 2 | | 分子名称: | S protein | | 著者 | Yuan, Y, Cao, D, Zhang, Y, Ma, J, Qi, J, Wang, Q, Lu, G, Wu, Y, Yan, J, Shi, Y, Zhang, X, Gao, G.F. | | 登録日 | 2017-02-15 | | 公開日 | 2017-05-03 | | 最終更新日 | 2017-05-24 | | 実験手法 | ELECTRON MICROSCOPY (4.2 Å) | | 主引用文献 | Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains
Nat Commun, 8, 2017
|
|
