8SXL
 
 | | RNA UU template binding to AMP monomer | | Descriptor: | ADENOSINE MONOPHOSPHATE, RNA (5'-R(*(TLN)P*(TLN)P*(LCA)P*(LCG)P*AP*CP*UP*UP*AP*AP*GP*UP*CP*U)-3') | | Authors: | Zhang, W, Dantsu, Y. | | Deposit date: | 2023-05-22 | | Release date: | 2023-05-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Insight into the structures of unusual base pairs in RNA complexes containing a primer/template/adenosine ligand.
Rsc Chem Biol, 4, 2023
|
|
8SWO
 
 | | GpppA dinucleotide ligand binding to RNA UC template | | Descriptor: | GUANOSINE-P3-ADENOSINE-5',5'-TRIPHOSPHATE, MAGNESIUM ION, RNA (5'-R(*(TLN)P*(LCC)P*(LCC)P*(LCG)P*AP*CP*UP*UP*AP*AP*GP*UP*CP*G*(GA3))-3') | | Authors: | Zhang, W, Dantsu, Y. | | Deposit date: | 2023-05-19 | | Release date: | 2023-05-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Insight into the structures of unusual base pairs in RNA complexes containing a primer/template/adenosine ligand.
Rsc Chem Biol, 4, 2023
|
|
8SY1
 
 | | RNA duplex bound with imidazolium bridged GA dinucleotide | | Descriptor: | RNA (5'-R(*(TLN)P*(LCC)P*(LCC)P*(LCG)P*AP*CP*UP*UP*AP*AP*GP*UP*CP*G*(GMA))-3'), [(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-[2-azanyl-3-[[(2~{R},3~{S},4~{R},5~{R})-5-(2-azanyl-6-oxidanylidene-1~{H}-purin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl]imidazol-1-yl]phosphinic acid | | Authors: | Zhang, W, Dantsu, Y. | | Deposit date: | 2023-05-24 | | Release date: | 2023-05-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Insight into the structures of unusual base pairs in RNA complexes containing a primer/template/adenosine ligand.
Rsc Chem Biol, 4, 2023
|
|
2L83
 
 | | A protein from Haloferax volcanii | | Descriptor: | Small archaeal modifier protein 1 | | Authors: | Zhang, W, Liao, S, Fan, K, Tu, X. | | Deposit date: | 2011-01-03 | | Release date: | 2012-01-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Ionic strength-dependent conformations of a ubiquitin-like small archaeal modifier protein (SAMP1) from Haloferax volcanii.
Protein Sci., 22, 2013
|
|
5ZMR
 
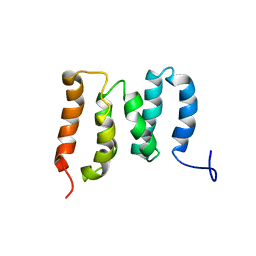 | | Solution Structure of the N-terminal Domain of the Yeast Rpn5 | | Descriptor: | 26S proteasome regulatory subunit RPN5 | | Authors: | Zhang, W, Zhao, C, Li, H, Hu, Y, Jin, C. | | Deposit date: | 2018-04-05 | | Release date: | 2018-09-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the N-terminal domain of proteasome lid subunit Rpn5
Biochem. Biophys. Res. Commun., 504, 2018
|
|
5TGP
 
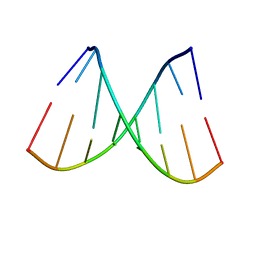 | | DNA 8mer containing two 2SeT modifications | | Descriptor: | DNA/RNA (5'-R(*G)-D(P*(2ST))-R(P*G)-D(P*(2ST))-R(P*AP*CP*AP*C)-3') | | Authors: | Zhang, W, Huang, Z. | | Deposit date: | 2016-09-28 | | Release date: | 2017-10-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | DNA 8mer containing two 2SeT modifications
To Be Published
|
|
8F27
 
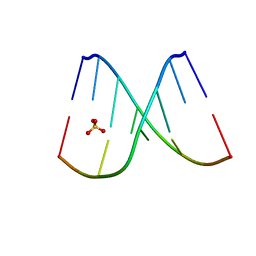 | | Mirror-image DNA containing 2'-OMe-L-uridine residue | | Descriptor: | Mirror-image DNA (5'-D(*(0DG)P*(0MU)P*(0DG)P*(0DT)P*(0DA)P*(0DC)P*(0DA)P*(0DC))-3'), SULFATE ION | | Authors: | Zhang, W, Dantsu, Y. | | Deposit date: | 2022-11-07 | | Release date: | 2023-04-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Derivatization of Mirror-Image l-Nucleic Acids with 2'-OMe Modification for Thermal and Structural Stabilization.
Chembiochem, 24, 2023
|
|
4ACS
 
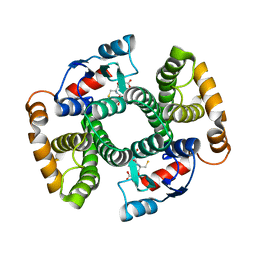 | | Crystal structure of mutant GST A2-2 with enhanced catalytic efficiency with azathioprine | | Descriptor: | GLUTATHIONE, GLUTATHIONE S-TRANSFERASE A2 | | Authors: | Zhang, W, Moden, O, Tars, K, Mannervik, B. | | Deposit date: | 2011-12-19 | | Release date: | 2011-12-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-based redesign of GST A2-2 for enhanced catalytic efficiency with azathioprine.
Chem.Biol., 19, 2012
|
|
6K0R
 
 | | Ruvbl1-Ruvbl2 with truncated domain II in complex with phosphorylated Cordycepin | | Descriptor: | 3'-DEOXYADENOSINE-5'-TRIPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Zhang, W, Chen, L, Li, W, Ju, D, Huang, N, Zhang, E. | | Deposit date: | 2019-05-07 | | Release date: | 2020-05-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Chemical perturbations reveal that RUVBL2 regulates the circadian phase in mammals.
Sci Transl Med, 12, 2020
|
|
8F5C
 
 | | Mirror-image DNA containing 2'-OMe-L-dC modification | | Descriptor: | DNA (5'-D(*(0DG))-R(P*(XE6))-D(P*(0DG)P*(0DT)P*(0DA)P*(0DC)P*(0DG)P*(0DC))-3'), MAGNESIUM ION | | Authors: | Zhang, W, Dantsu, Y. | | Deposit date: | 2022-11-13 | | Release date: | 2023-09-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Synthesis and Structural Characterization of 2'-Deoxy-2'-Methoxy-L-Cytidine Nucleic Acids
Chemistryselect, 2023
|
|
3CMI
 
 | |
6U6J
 
 | | RNA-monomer complex containing pyrophosphate linkage | | Descriptor: | 5'-O-[(R)-(2-amino-1H-imidazol-1-yl)(hydroxy)phosphoryl]guanosine, RNA (5'-R(*(LCC)P*(LCC)P*(LCC)P*(LCG)P*AP*CP*UP*UP*AP*AP*GP*UP*CP*G*(DPG))-3') | | Authors: | Zhang, W, Szostak, J.W, Giurgiu, C, Wright, T. | | Deposit date: | 2019-08-29 | | Release date: | 2019-11-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Prebiotically Plausible "Patching" of RNA Backbone Cleavage through a 3'-5' Pyrophosphate Linkage.
J.Am.Chem.Soc., 141, 2019
|
|
8SPH
 
 | | Crystal structure of chimeric omicron RBD (strain XBB.1) complexed with human ACE2 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, W, Shi, K, Aihara, H, Li, F. | | Deposit date: | 2023-05-03 | | Release date: | 2023-08-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Structural evolution of SARS-CoV-2 omicron in human receptor recognition.
J.Virol., 97, 2023
|
|
8SPI
 
 | | Crystal structure of chimeric omicron RBD (strain XBB.1.5) complexed with human ACE2 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, W, Shi, K, Aihara, H, Li, F. | | Deposit date: | 2023-05-03 | | Release date: | 2023-08-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | Structural evolution of SARS-CoV-2 omicron in human receptor recognition.
J.Virol., 97, 2023
|
|
6UR4
 
 | |
6UR9
 
 | | DNA polymerase I Large Fragment from Bacillus stearothermophilus with DNA template, dideoxy primer, 3'-amino-ddGTP (nGTP), and Ca2+ | | Descriptor: | 3'-amino-2',3'-dideoxyguanosine 5'-(tetrahydrogen triphosphate), CALCIUM ION, DNA (5'-D(*CP*AP*CP*GP*CP*TP*GP*AP*TP*CP*GP*CP*A)-3'), ... | | Authors: | Zhang, W, Lelyveld, V.S, Szostak, J.W. | | Deposit date: | 2019-10-22 | | Release date: | 2020-03-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Synthesis of phosphoramidate-linked DNA by a modified DNA polymerase.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6US5
 
 | | DNA polymerase I Large Fragment from Bacillus stearothermophilus with DNA template, 3'-amino primer, dGpNHpp analog, and Mn2+ | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]guanosine, DNA (5'-D(*CP*AP*CP*GP*CP*TP*GP*AP*TP*CP*GP*CP*A)-3'), DNA (5'-D(*GP*CP*GP*AP*TP*CP*AP*GP*(C42))-3'), ... | | Authors: | Zhang, W, Lelyveld, V.S, Szostak, J.W. | | Deposit date: | 2019-10-24 | | Release date: | 2020-03-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Synthesis of phosphoramidate-linked DNA by a modified DNA polymerase.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6UR2
 
 | |
3HCK
 
 | |
7UFK
 
 | | Crystal structure of chimeric omicron RBD (strain BA.2) complexed with human ACE2 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, W, Shi, K, Geng, Q, Ye, G, Aihara, H, Li, F. | | Deposit date: | 2022-03-22 | | Release date: | 2022-10-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Structural basis for mouse receptor recognition by SARS-CoV-2 omicron variant.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7UFL
 
 | | Crystal structure of chimeric omicron RBD (strain BA.2) complexed with chimeric mouse ACE2 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, W, Shi, K, Geng, Q, Ye, G, Aihara, H, Li, F. | | Deposit date: | 2022-03-22 | | Release date: | 2022-10-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Structural basis for mouse receptor recognition by SARS-CoV-2 omicron variant.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
5GQ0
 
 | |
5GQT
 
 | |
5HBY
 
 | | RNA primer-template complex with 2-methylimidazole-activated monomer analogue-3 binding sites | | Descriptor: | MAGNESIUM ION, RNA (5'-R(*(LCC)P*(LCC)P*(LCC)P*(LCG)P*AP*CP*UP*UP*AP*AP*GP*UP*C)-3'), [(2~{R},3~{S},4~{R},5~{R})-5-(2-azanyl-6-oxidanylidene-1~{H}-purin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-(3-methyl-1~{H}-pyrazol-4-yl)phosphinic acid | | Authors: | Zhang, W, Tam, C.P, Wang, J, Szostak, J.W. | | Deposit date: | 2016-01-03 | | Release date: | 2016-12-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Unusual Base-Pairing Interactions in Monomer-Template Complexes.
ACS Cent Sci, 2, 2016
|
|
5HBW
 
 | | RNA primer-template complex with 2-methylimidazole-activated monomer analogue | | Descriptor: | RNA (5'-R(*(LCC)P*(TLN)P*(LCG)P*UP*AP*CP*A)-3'), [(2~{R},3~{S},4~{R},5~{R})-5-(2-azanyl-6-oxidanylidene-1~{H}-purin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-(3-methyl-1~{H}-pyrazol-4-yl)phosphinic acid | | Authors: | Zhang, W, Tam, C.P, Szostak, J.W. | | Deposit date: | 2016-01-03 | | Release date: | 2016-12-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Unusual Base-Pairing Interactions in Monomer-Template Complexes.
ACS Cent Sci, 2, 2016
|
|
