4Y5E
 
 | | Endothiapepsin in complex with fragment 268 | | Descriptor: | (2R)-N-(trans-4-methylcyclohexyl)tetrahydrofuran-2-carboxamide, ACETATE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Wang, X, Heine, A, Klebe, G. | | Deposit date: | 2015-02-11 | | Release date: | 2016-03-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.116 Å) | | Cite: | Crystallographic Fragment Sreening of an Entire Library
to be published
|
|
4Y58
 
 | |
4Y5K
 
 | | Endothiapepsin in complex with fragment 274 | | Descriptor: | (1S)-2-amino-1-(4-fluorophenyl)ethanol, ACETATE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Wang, X, Heine, A, Klebe, G. | | Deposit date: | 2015-02-11 | | Release date: | 2016-03-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.439 Å) | | Cite: | Crystallographic Fragment Sreening of an Entire Library
to be published
|
|
7XNQ
 
 | | SARS-CoV-2 Omicron BA.4 variant spike | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Wang, X, Wang, L. | | Deposit date: | 2022-04-29 | | Release date: | 2022-07-13 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | BA.2.12.1, BA.4 and BA.5 escape antibodies elicited by Omicron infection.
Nature, 608, 2022
|
|
7XNS
 
 | | SARS-CoV-2 Omicron BA.2.12.1 variant spike | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Wang, X, Wang, L. | | Deposit date: | 2022-04-29 | | Release date: | 2022-07-13 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | BA.2.12.1, BA.4 and BA.5 escape antibodies elicited by Omicron infection.
Nature, 608, 2022
|
|
7XNR
 
 | | SARS-CoV-2 Omicron BA.2.13 variant spike | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Wang, X, Wang, L. | | Deposit date: | 2022-04-29 | | Release date: | 2022-07-13 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | BA.2.12.1, BA.4 and BA.5 escape antibodies elicited by Omicron infection.
Nature, 608, 2022
|
|
4Y5G
 
 | |
8VOI
 
 | |
8VOH
 
 | |
3HBJ
 
 | | Structure of UGT78G1 complexed with UDP | | Descriptor: | Flavonoid 3-O-glucosyltransferase, URIDINE-5'-DIPHOSPHATE | | Authors: | Wang, X, Modolo, L, Li, L, Dixon, R. | | Deposit date: | 2009-05-04 | | Release date: | 2009-09-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of glycosyltransferase UGT78G1 reveal the molecular basis for glycosylation and deglycosylation of (iso)flavonoids.
J.Mol.Biol., 392, 2009
|
|
2B5I
 
 | | cytokine receptor complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cytokine receptor common gamma chain, ... | | Authors: | Wang, X, Rickert, M, Garcia, K.C. | | Deposit date: | 2005-09-28 | | Release date: | 2005-11-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the quaternary complex of interleukin-2 with its alpha, beta, and gammac receptors.
Science, 310, 2005
|
|
8JUE
 
 | | Crystal structure of glutaminase C in complex with compound 11 | | Descriptor: | 2-(3-phenoxyphenyl)-N-[5-[[(3R)-1-pyridazin-3-ylpyrrolidin-3-yl]amino]-1,3,4-thiadiazol-2-yl]ethanamide, Glutaminase kidney isoform, mitochondrial | | Authors: | Wang, X, Hanyu, S, Tingting, D. | | Deposit date: | 2023-06-26 | | Release date: | 2023-10-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Targeting the Subpocket Enables the Discovery of Thiadiazole-Pyridazine Derivatives as Glutaminase C Inhibitors.
Acs Med.Chem.Lett., 14, 2023
|
|
8JUB
 
 | | Crystal structure of glutaminase C in complex with compound 27 | | Descriptor: | 3-[2-oxidanylidene-2-[[5-[[(3R)-1-pyridazin-3-ylpyrrolidin-3-yl]amino]-1,3,4-thiadiazol-2-yl]amino]ethyl]benzoic acid, Glutaminase kidney isoform, mitochondrial | | Authors: | Wang, X, Hanyu, S, Tingting, D. | | Deposit date: | 2023-06-26 | | Release date: | 2023-10-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Targeting the Subpocket Enables the Discovery of Thiadiazole-Pyridazine Derivatives as Glutaminase C Inhibitors.
Acs Med.Chem.Lett., 14, 2023
|
|
5XEX
 
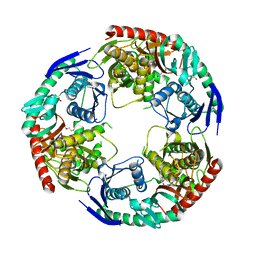 | | Crystal structure of S.aureus PNPase catalytic domain | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, PYROPHOSPHATE, ... | | Authors: | Wang, X, Zhang, X, Zang, J. | | Deposit date: | 2017-04-06 | | Release date: | 2017-10-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Enolase binds to RnpA in competition with PNPase in Staphylococcus aureus
FEBS Lett., 591, 2017
|
|
6XS6
 
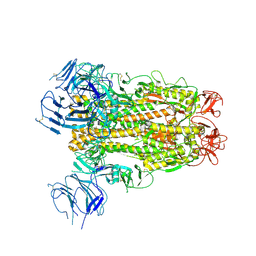 | | SARS-CoV-2 Spike D614G variant, minus RBD | | Descriptor: | Spike glycoprotein | | Authors: | Wang, X, Egri, S.B, Dudkina, N, Luban, J, Shen, K. | | Deposit date: | 2020-07-15 | | Release date: | 2020-07-22 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural and Functional Analysis of the D614G SARS-CoV-2 Spike Protein Variant.
Cell, 183, 2020
|
|
7MIZ
 
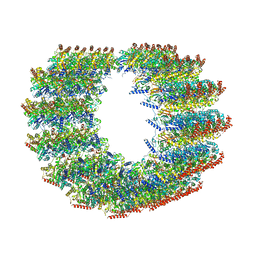 | | Atomic structure of cortical microtubule from Toxoplasma gondii | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Wang, X, Brown, A, Sibley, L.D, Zhang, R. | | Deposit date: | 2021-04-18 | | Release date: | 2021-06-02 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structure of cortical microtubules from human parasite Toxoplasma gondii identifies their microtubule inner proteins.
Nat Commun, 12, 2021
|
|
6M0J
 
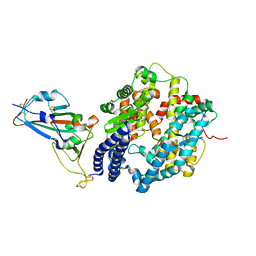 | | Crystal structure of SARS-CoV-2 spike receptor-binding domain bound with ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, CHLORIDE ION, ... | | Authors: | Wang, X, Lan, J, Ge, J, Yu, J, Shan, S. | | Deposit date: | 2020-02-21 | | Release date: | 2020-03-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor.
Nature, 581, 2020
|
|
7LGI
 
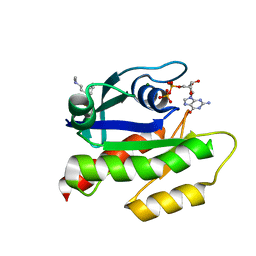 | | The haddock model of GDP KRas in complex with promazine using chemical shift perturbations and intermolecular NOEs | | Descriptor: | GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Wang, X, Gorfe, A.A, Putkey, J.A. | | Deposit date: | 2021-01-20 | | Release date: | 2021-07-21 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Antipsychotic phenothiazine drugs bind to KRAS in vitro.
J.Biomol.Nmr, 75, 2021
|
|
7MQU
 
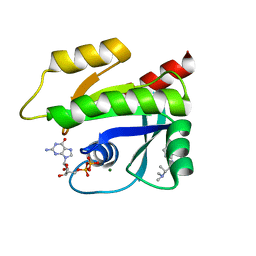 | | The haddock model of GDP KRas in complex with promethazine using NMR chemical shift perturbations | | Descriptor: | (2R)-N,N-dimethyl-1-(10H-phenothiazin-10-yl)propan-2-amine, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Wang, X, Gorfe, A.A, Putkey, J.P. | | Deposit date: | 2021-05-06 | | Release date: | 2022-05-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Antipsychotic phenothiazine drugs bind to KRAS in vitro.
J.Biomol.Nmr, 75, 2021
|
|
4EGT
 
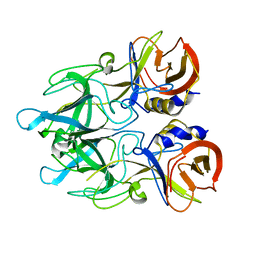 | | Crystal structure of major capsid protein P domain from rabbit hemorrhagic disease virus | | Descriptor: | Major capsid protein VP60 | | Authors: | Wang, X, Xu, F, Zhang, K, Zhai, Y, Sun, F. | | Deposit date: | 2012-04-01 | | Release date: | 2013-01-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Atomic model of rabbit hemorrhagic disease virus by cryo-electron microscopy and crystallography.
Plos Pathog., 9, 2013
|
|
3HBF
 
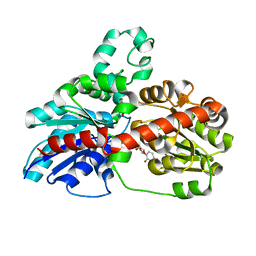 | | Structure of UGT78G1 complexed with myricetin and UDP | | Descriptor: | 3,5,7-TRIHYDROXY-2-(3,4,5-TRIHYDROXYPHENYL)-4H-CHROMEN-4-ONE, Flavonoid 3-O-glucosyltransferase, URIDINE-5'-DIPHOSPHATE | | Authors: | Wang, X, Modolo, L, Li, L, Dixon, R. | | Deposit date: | 2009-05-04 | | Release date: | 2009-09-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of glycosyltransferase UGT78G1 reveal the molecular basis for glycosylation and deglycosylation of (iso)flavonoids.
J.Mol.Biol., 392, 2009
|
|
5HLY
 
 | |
7XSA
 
 | |
7XSC
 
 | |
7XSB
 
 | |
