7YW7
 
 | |
7YW8
 
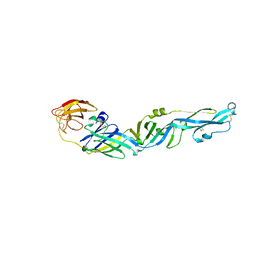 | |
7WHH
 
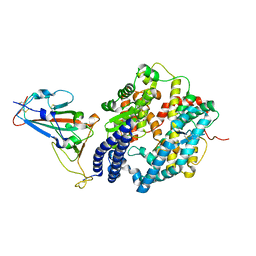 | | Crystal structure of SARS-CoV-2 omicron RBD and human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Processed angiotensin-converting enzyme 2, ... | | Authors: | Wang, X.Q, Lan, J. | | Deposit date: | 2021-12-30 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of SARS-CoV-2 omicron RBD and human ACE2
To Be Published
|
|
7Y96
 
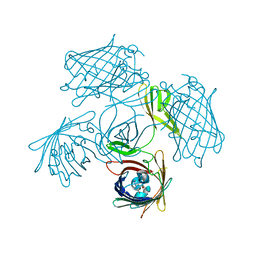 | |
7FC3
 
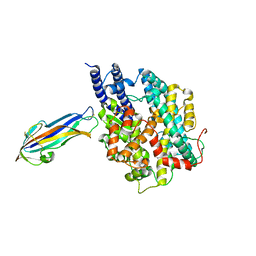 | | structure of NL63 receptor-binding domain complexed with horse ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, ... | | Authors: | Wang, X.Q, Ge, J.W, Lan, J. | | Deposit date: | 2021-07-13 | | Release date: | 2021-09-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Structural insights into the binding of SARS-CoV-2, SARS-CoV, and hCoV-NL63 spike receptor-binding domain to horse ACE2.
Structure, 30, 2022
|
|
2L9H
 
 | | Oligomeric Structure of the Chemokine CCL5/RANTES from NMR, MS, and SAXS Data | | Descriptor: | C-C motif chemokine 5 | | Authors: | Wang, X, Watson, C.M, Sharp, J.S, Handel, T.M, Prestegard, J.H. | | Deposit date: | 2011-02-09 | | Release date: | 2011-06-22 | | Last modified: | 2011-08-24 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | Oligomeric Structure of the Chemokine CCL5/RANTES from NMR, MS, and SAXS Data.
Structure, 19, 2011
|
|
7D1L
 
 | | complex structure of two RRM domains | | Descriptor: | Embryonic developmental protein tofu-6, Uncharacterized protein | | Authors: | Wang, X, Liao, S, Xu, C. | | Deposit date: | 2020-09-14 | | Release date: | 2021-08-25 | | Last modified: | 2021-10-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Molecular basis for PICS-mediated piRNA biogenesis and cell division.
Nat Commun, 12, 2021
|
|
7D2Y
 
 | | complex of two RRM domains | | Descriptor: | Embryonic developmental protein tofu-6, RRM2, SULFATE ION | | Authors: | Wang, X, Liao, S, Xu, C. | | Deposit date: | 2020-09-17 | | Release date: | 2021-08-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Molecular basis for PICS-mediated piRNA biogenesis and cell division.
Nat Commun, 12, 2021
|
|
7EJS
 
 | | Structure of ERH-2 bound to PICS-1 | | Descriptor: | Enhancer of rudimentary homolog 2,Protein pid-3 | | Authors: | Wang, X, Xu, C. | | Deposit date: | 2021-04-02 | | Release date: | 2021-08-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.387 Å) | | Cite: | Molecular basis for PICS-mediated piRNA biogenesis and cell division.
Nat Commun, 12, 2021
|
|
6M16
 
 | | Cryo-EM structures of SADS-CoV spike glycoproteins | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Wang, X, Yu, J, Qiao, S, Guo, R. | | Deposit date: | 2020-02-24 | | Release date: | 2020-05-27 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Cryo-EM structures of HKU2 and SADS-CoV spike glycoproteins provide insights into coronavirus evolution.
Nat Commun, 11, 2020
|
|
7EJO
 
 | | Structure of ERH-2 bound to TOST-1 | | Descriptor: | Enhancer of rudimentary homolog 2, Enhancer of rudimentary homolog 2,Protein tost-1 | | Authors: | Wang, X, Xu, C. | | Deposit date: | 2021-04-02 | | Release date: | 2021-08-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.191 Å) | | Cite: | Molecular basis for PICS-mediated piRNA biogenesis and cell division.
Nat Commun, 12, 2021
|
|
7WLY
 
 | | Cryo-EM structure of the Omicron S in complex with 35B5 Fab(1 down- and 2 up RBDs) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of 35B5 Fab, ... | | Authors: | Wang, X, Zhu, Y. | | Deposit date: | 2022-01-14 | | Release date: | 2022-05-25 | | Last modified: | 2022-06-22 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | 35B5 antibody potently neutralizes SARS-CoV-2 Omicron by disrupting the N-glycan switch via a conserved spike epitope.
Cell Host Microbe, 30, 2022
|
|
7EGR
 
 | | Co-crystal structure of Ac-AChBPP in complex with RgIA | | Descriptor: | MAGNESIUM ION, RgIA, Soluble acetylcholine receptor | | Authors: | Wang, X.Q, Pan, S, Fan, Y.X, Xue, Y, Zhu, X.P, Luo, S.L. | | Deposit date: | 2021-03-26 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.503 Å) | | Cite: | Co-crystal structure of Ac-AChBPP in complex with RgIA
To Be Published
|
|
7Y9B
 
 | | Crystal structure of the membrane (M) protein of a SARS-COV-2-related coronavirus | | Descriptor: | 3,6,9,12,15-PENTAOXATRICOSAN-1-OL, Membrane protein | | Authors: | Wang, X, Sun, Z, Zhou, X. | | Deposit date: | 2022-06-24 | | Release date: | 2022-08-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.214 Å) | | Cite: | Crystal structure of the membrane (M) protein from a bat betacoronavirus.
Pnas Nexus, 2, 2023
|
|
7BIK
 
 | | Crystal structure of YTHDF2 in complex with m6Am | | Descriptor: | (2~{R},3~{S},4~{R},5~{R})-2-(hydroxymethyl)-4-methoxy-5-[6-(methylamino)purin-9-yl]oxolan-3-ol, GLYCEROL, SULFATE ION, ... | | Authors: | Wang, X, Caflisch, A. | | Deposit date: | 2021-01-12 | | Release date: | 2021-11-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of YTHDF2 in complex with m6Am
To Be Published
|
|
7WLZ
 
 | |
7CYG
 
 | | Crystal structure of a cysteine-pair mutant (Y113C-P190C) of a bacterial bile acid transporter before disulfide bond formation | | Descriptor: | Transporter, sodium/bile acid symporter family | | Authors: | Wang, X, Lyu, Y, Ji, Y, Sun, Z, Zhou, X. | | Deposit date: | 2020-09-03 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.198 Å) | | Cite: | An engineered disulfide bridge traps and validates an outward-facing conformation in a bile acid transporter.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7CYK
 
 | | Crystal structure of a second cysteine-pair mutant (V110C-I197C) of a bacterial bile acid transporter before disulfide bond formation | | Descriptor: | MERCURY (II) ION, Transporter, sodium/bile acid symporter family | | Authors: | Wang, X, Lyu, Y, Ji, Y, Sun, Z, Zhou, X. | | Deposit date: | 2020-09-03 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.785 Å) | | Cite: | An engineered disulfide bridge traps and validates an outward-facing conformation in a bile acid transporter.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
6M15
 
 | | Cryo-EM structures of HKU2 spike glycoproteins | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Wang, X, Yu, J, Qiao, S, Guo, R. | | Deposit date: | 2020-02-24 | | Release date: | 2020-05-27 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (2.38 Å) | | Cite: | Cryo-EM structures of HKU2 and SADS-CoV spike glycoproteins provide insights into coronavirus evolution.
Nat Commun, 11, 2020
|
|
7WM0
 
 | |
7X39
 
 | | Structure of CIZ1 bound ERH | | Descriptor: | Enhancer of rudimentary homolog,Cip1-interacting zinc finger protein | | Authors: | Wang, X, Xu, C. | | Deposit date: | 2022-02-28 | | Release date: | 2022-08-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Molecular basis for the recognition of CIZ1 by ERH.
Febs J., 290, 2023
|
|
7CVN
 
 | | The N-arylsulfonyl-indole-2-carboxamide-based inhibitors against fructose-1,6-bisphosphatase | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, 4-(3-acetamidophenyl)-N-(4-methoxyphenyl)sulfonyl-7-nitro-1H-indole-2-carboxamide, Fructose-1,6-bisphosphatase 1 | | Authors: | Wang, X, Zhou, J, Xu, B. | | Deposit date: | 2020-08-26 | | Release date: | 2020-09-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Design,synthesis,biological evaluation and binding mode analysis of 7-nitro-indole-N-acylarylsulfonamide-based fructose-1,6-bisphosphatase inhibitors
Chinese journal of medicinal chemistry, 30, 2020
|
|
2LQU
 
 | |
2K2R
 
 | | The NMR structure of alpha-parvin CH2/paxillin LD1 complex | | Descriptor: | Alpha-parvin, Paxillin | | Authors: | Wang, X, Fukuda, K, Byeon, I, Velyvis, A, Wu, C, Gronenborn, A, Qin, J. | | Deposit date: | 2008-04-10 | | Release date: | 2008-05-27 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The Structure of {alpha}-Parvin CH2-Paxillin LD1 Complex Reveals a Novel Modular Recognition for Focal Adhesion Assembly.
J.Biol.Chem., 283, 2008
|
|
2MTD
 
 | |
