3UVY
 
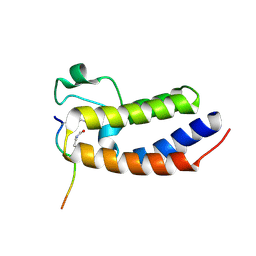 | | Crystal Structure of the first bromodomain of human BRD4 in complex with a diacetylated histone 4 peptide (H4K16acK20ac) | | Descriptor: | Bromodomain-containing protein 4, Histone H4 | | Authors: | Filippakopoulos, P, Felletar, I, Picaud, S, Keates, T, Muniz, J, Gileadi, O, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-11-30 | | Release date: | 2012-01-18 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Histone recognition and large-scale structural analysis of the human bromodomain family.
Cell(Cambridge,Mass.), 149, 2012
|
|
4USD
 
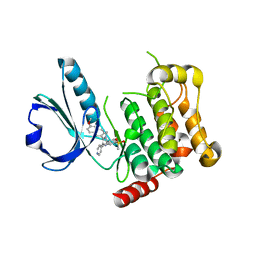 | | Human STK10 (LOK) with SB-633825 | | Descriptor: | 4-{5-(6-methoxynaphthalen-2-yl)-1-methyl-2-[2-methyl-4-(methylsulfonyl)phenyl]-1H-imidazol-4-yl}pyridine, SERINE/THREONINE-PROTEIN KINASE 10 | | Authors: | Elkins, J.M, Salah, E, Szklarz, M, von Delft, F, Canning, P, Raynor, J, Bountra, C, Edwards, A.M, Knapp, S. | | Deposit date: | 2014-07-07 | | Release date: | 2015-07-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Comprehensive Characterization of the Published Kinase Inhibitor Set.
Nat.Biotechnol., 34, 2016
|
|
3UVD
 
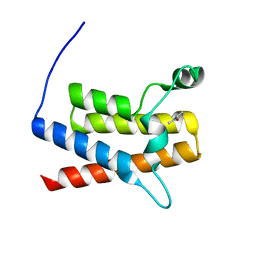 | | Crystal Structure of the bromodomain of human Transcription activator BRG1 (SMARCA4) in complex with N-Methyl-2-pyrrolidone | | Descriptor: | 1-methylpyrrolidin-2-one, Transcription activator BRG1 | | Authors: | Filippakopoulos, P, Felletar, I, Picaud, S, Keates, T, Muniz, J, Gileadi, O, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-11-29 | | Release date: | 2012-01-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Histone recognition and large-scale structural analysis of the human bromodomain family.
Cell(Cambridge,Mass.), 149, 2012
|
|
5L6W
 
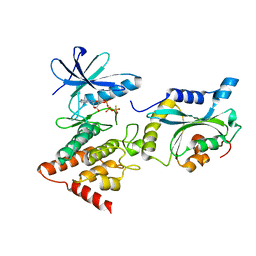 | | Structure Of the LIMK1-ATPgammaS-CFL1 Complex | | Descriptor: | Cofilin-1, LIM domain kinase 1, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | Authors: | Salah, E, Mathea, S, Oerum, S, Newman, J.A, Tallant, C, Adamson, R, Canning, P, Beltrami, A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Bullock, A.N. | | Deposit date: | 2016-05-31 | | Release date: | 2016-06-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Structure Of the LIMK1-ATPgammaS-CFL1 Complex
To Be Published
|
|
5SLA
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NSP14 in complex with Z1003207278 | | Descriptor: | 1-cyclohexyl-N-methylmethanesulfonamide, PHOSPHATE ION, Proofreading exoribonuclease nsp14, ... | | Authors: | Imprachim, N, Yosaatmadja, Y, von-Delft, F, Bountra, C, Gileadi, O, Newman, J.A. | | Deposit date: | 2022-03-03 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5SLP
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NSP14 in complex with Z373768898 | | Descriptor: | N-(1-ethyl-1H-pyrazol-4-yl)cyclopentanecarboxamide, PHOSPHATE ION, Proofreading exoribonuclease nsp14, ... | | Authors: | Imprachim, N, Yosaatmadja, Y, von-Delft, F, Bountra, C, Gileadi, O, Newman, J.A. | | Deposit date: | 2022-03-03 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.819 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5SL6
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NSP14 in complex with Z256709556 | | Descriptor: | 3-methylthiophene-2-carboxylic acid, PHOSPHATE ION, Proofreading exoribonuclease nsp14, ... | | Authors: | Imprachim, N, Yosaatmadja, Y, von-Delft, F, Bountra, C, Gileadi, O, Newman, J.A. | | Deposit date: | 2022-03-03 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.289 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5SLK
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NSP14 in complex with Z1354370680 | | Descriptor: | 2-[(5-chloro-3-fluoropyridin-2-yl)(methyl)amino]ethan-1-ol, PHOSPHATE ION, Proofreading exoribonuclease nsp14, ... | | Authors: | Imprachim, N, Yosaatmadja, Y, von-Delft, F, Bountra, C, Gileadi, O, Newman, J.A. | | Deposit date: | 2022-03-03 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5SM4
 
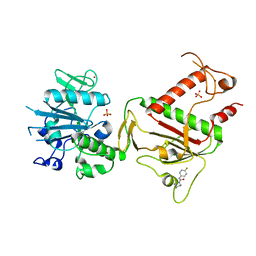 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NSP14 in complex with Z2856434944 | | Descriptor: | PHOSPHATE ION, Proofreading exoribonuclease nsp14, ZINC ION, ... | | Authors: | Imprachim, N, Yosaatmadja, Y, von-Delft, F, Bountra, C, Gileadi, O, Newman, J.A. | | Deposit date: | 2022-03-03 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5SLS
 
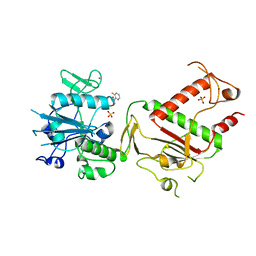 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NSP14 in complex with Z1373445602 | | Descriptor: | 4-(3-fluoranylpyridin-2-yl)-1-methyl-piperazin-2-one, PHOSPHATE ION, Proofreading exoribonuclease nsp14, ... | | Authors: | Imprachim, N, Yosaatmadja, Y, von-Delft, F, Bountra, C, Gileadi, O, Newman, J.A. | | Deposit date: | 2022-03-03 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5SMB
 
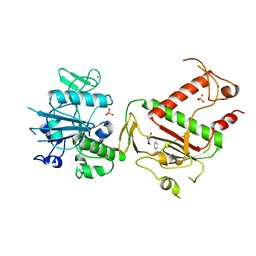 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NSP14 in complex with Z419995480 | | Descriptor: | 1-(morpholin-4-yl)-4-phenylbutan-1-one, PHOSPHATE ION, Proofreading exoribonuclease nsp14, ... | | Authors: | Imprachim, N, Yosaatmadja, Y, von-Delft, F, Bountra, C, Gileadi, O, Newman, J.A. | | Deposit date: | 2022-03-03 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.181 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5SL1
 
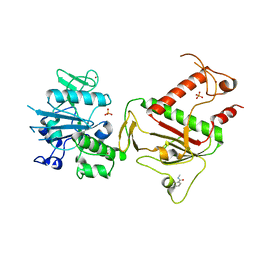 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NSP14 in complex with Z1273312153 | | Descriptor: | N-methyl-1H-indole-7-carboxamide, PHOSPHATE ION, Proofreading exoribonuclease nsp14, ... | | Authors: | Imprachim, N, Yosaatmadja, Y, von-Delft, F, Bountra, C, Gileadi, O, Newman, J.A. | | Deposit date: | 2022-03-03 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.383 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5SLG
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NSP14 in complex with Z32400357 | | Descriptor: | 1-(3-methylbenzene-1-carbonyl)piperidine-4-carboxamide, PHOSPHATE ION, Proofreading exoribonuclease nsp14, ... | | Authors: | Imprachim, N, Yosaatmadja, Y, von-Delft, F, Bountra, C, Gileadi, O, Newman, J.A. | | Deposit date: | 2022-03-03 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5SKY
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NSP14 in complex with Z466628048 | | Descriptor: | N-[(4-methyl-1,3-thiazol-2-yl)methyl]-1H-pyrazole-5-carboxamide, PHOSPHATE ION, Proofreading exoribonuclease nsp14, ... | | Authors: | Imprachim, N, Yosaatmadja, Y, von-Delft, F, Bountra, C, Gileadi, O, Newman, J.A. | | Deposit date: | 2022-03-03 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5SLD
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NSP14 in complex with Z1246465616 | | Descriptor: | (2R)-3-(3,5-dimethyl-1,2-oxazol-4-yl)-N,N,2-trimethylpropanamide, PHOSPHATE ION, Proofreading exoribonuclease nsp14, ... | | Authors: | Imprachim, N, Yosaatmadja, Y, von-Delft, F, Bountra, C, Gileadi, O, Newman, J.A. | | Deposit date: | 2022-03-03 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.581 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5SLY
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NSP14 in complex with Z1526504764 | | Descriptor: | 1-(1-ethyl-1H-pyrazol-5-yl)-N-methylmethanamine, PHOSPHATE ION, Proofreading exoribonuclease nsp14, ... | | Authors: | Imprachim, N, Yosaatmadja, Y, von-Delft, F, Bountra, C, Gileadi, O, Newman, J.A. | | Deposit date: | 2022-03-03 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.018 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5SMC
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NSP14 in complex with Z2033637875 | | Descriptor: | N~2~-(4-cyano-3-methyl-1,2-thiazol-5-yl)-N~2~-methylglycinamide, PHOSPHATE ION, Proofreading exoribonuclease nsp14, ... | | Authors: | Imprachim, N, Yosaatmadja, Y, von-Delft, F, Bountra, C, Gileadi, O, Newman, J.A. | | Deposit date: | 2022-03-03 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5SLT
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NSP14 in complex with Z1816233707 | | Descriptor: | 6-(methylcarbamoyl)pyridine-2-carboxylic acid, PHOSPHATE ION, Proofreading exoribonuclease nsp14, ... | | Authors: | Imprachim, N, Yosaatmadja, Y, von-Delft, F, Bountra, C, Gileadi, O, Newman, J.A. | | Deposit date: | 2022-03-03 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5SM7
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NSP14 in complex with Z1247413608 | | Descriptor: | 1-(methanesulfonyl)piperidin-4-ol, PHOSPHATE ION, Proofreading exoribonuclease nsp14, ... | | Authors: | Imprachim, N, Yosaatmadja, Y, von-Delft, F, Bountra, C, Gileadi, O, Newman, J.A. | | Deposit date: | 2022-03-03 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5SL4
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NSP14 in complex with Z383202616 | | Descriptor: | N-(1H-indazol-6-yl)acetamide, PHOSPHATE ION, Proofreading exoribonuclease nsp14, ... | | Authors: | Imprachim, N, Yosaatmadja, Y, von-Delft, F, Bountra, C, Gileadi, O, Newman, J.A. | | Deposit date: | 2022-03-03 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.936 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5SLF
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NSP14 in complex with Z198195770 | | Descriptor: | N-[3-(carbamoylamino)phenyl]acetamide, PHOSPHATE ION, Proofreading exoribonuclease nsp14, ... | | Authors: | Imprachim, N, Yosaatmadja, Y, von-Delft, F, Bountra, C, Gileadi, O, Newman, J.A. | | Deposit date: | 2022-03-03 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5SL0
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NSP14 in complex with Z57260516 | | Descriptor: | 2-methoxy-~{N}-(2,4,6-trimethylphenyl)ethanamide, PHOSPHATE ION, Proofreading exoribonuclease nsp14, ... | | Authors: | Imprachim, N, Yosaatmadja, Y, von-Delft, F, Bountra, C, Gileadi, O, Newman, J.A. | | Deposit date: | 2022-03-03 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5SL5
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NSP14 in complex with Z32014663 | | Descriptor: | N,N,2,3-tetramethylbenzamide, PHOSPHATE ION, Proofreading exoribonuclease nsp14, ... | | Authors: | Imprachim, N, Yosaatmadja, Y, von-Delft, F, Bountra, C, Gileadi, O, Newman, J.A. | | Deposit date: | 2022-03-03 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5SLE
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NSP14 in complex with Z56880342 | | Descriptor: | N-ethyl-N'-(5-methyl-1,2-oxazol-3-yl)urea, PHOSPHATE ION, Proofreading exoribonuclease nsp14, ... | | Authors: | Imprachim, N, Yosaatmadja, Y, von-Delft, F, Bountra, C, Gileadi, O, Newman, J.A. | | Deposit date: | 2022-03-03 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.009 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5SLL
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 NSP14 in complex with Z54615640 | | Descriptor: | N-[(3R)-3-methyl-1,1-dioxo-1lambda~6~-thiolan-3-yl]cyclopropanecarboxamide, PHOSPHATE ION, Proofreading exoribonuclease nsp14, ... | | Authors: | Imprachim, N, Yosaatmadja, Y, von-Delft, F, Bountra, C, Gileadi, O, Newman, J.A. | | Deposit date: | 2022-03-03 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.807 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
