5SDR
 
 | | PanDDA analysis group deposition -- Crystal Structure of Porphyromonas gingivalis in complex with Z1273312153 | | Descriptor: | Asp/Glu-specific dipeptidyl-peptidase, CHLORIDE ION, N-methyl-1H-indole-7-carboxamide | | Authors: | Tham, C.T, Coker, J.A, Krojer, T, Foster, W.R, Koekemoer, L, Douangamath, A, Talon, R, Fearon, D, von Delft, F, Yue, W.W, Bountra, C, Bezerra, G.A. | | Deposit date: | 2022-01-20 | | Release date: | 2022-02-09 | | Method: | X-RAY DIFFRACTION (2.077 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5SDI
 
 | | PanDDA analysis group deposition -- Crystal Structure of Porphyromonas gingivalis in complex with Z44592329 | | Descriptor: | Asp/Glu-specific dipeptidyl-peptidase, CHLORIDE ION, N-phenyl-N'-pyridin-3-ylurea | | Authors: | Tham, C.T, Coker, J.A, Krojer, T, Foster, W.R, Koekemoer, L, Douangamath, A, Talon, R, Fearon, D, von Delft, F, Yue, W.W, Bountra, C, Bezerra, G.A. | | Deposit date: | 2022-01-20 | | Release date: | 2022-02-09 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5SDF
 
 | | PanDDA analysis group deposition -- Crystal Structure of Porphyromonas gingivalis in complex with Z2856434834 | | Descriptor: | Asp/Glu-specific dipeptidyl-peptidase, CHLORIDE ION, N-(2,3-dimethylphenyl)-2-(morpholin-4-yl)acetamide | | Authors: | Tham, C.T, Coker, J.A, Krojer, T, Foster, W.R, Koekemoer, L, Douangamath, A, Talon, R, Fearon, D, von Delft, F, Yue, W.W, Bountra, C, Bezerra, G.A. | | Deposit date: | 2022-01-20 | | Release date: | 2022-02-09 | | Method: | X-RAY DIFFRACTION (1.877 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5SDN
 
 | | PanDDA analysis group deposition -- Crystal Structure of Porphyromonas gingivalis in complex with Z437584380 | | Descriptor: | (4-chlorophenyl)(thiomorpholin-4-yl)methanone, Asp/Glu-specific dipeptidyl-peptidase, CHLORIDE ION | | Authors: | Tham, C.T, Coker, J.A, Krojer, T, Foster, W.R, Koekemoer, L, Douangamath, A, Talon, R, Fearon, D, von Delft, F, Yue, W.W, Bountra, C, Bezerra, G.A. | | Deposit date: | 2022-01-20 | | Release date: | 2022-02-09 | | Method: | X-RAY DIFFRACTION (2.023 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5SDD
 
 | | PanDDA analysis group deposition -- Crystal Structure of Porphyromonas gingivalis in complex with Z2856434879 | | Descriptor: | 2-[(4-methyl-1H-imidazol-5-yl)methyl]-1,2,3,4-tetrahydroisoquinoline, Asp/Glu-specific dipeptidyl-peptidase, CHLORIDE ION | | Authors: | Tham, C.T, Coker, J.A, Krojer, T, Foster, W.R, Koekemoer, L, Douangamath, A, Talon, R, Fearon, D, von Delft, F, Yue, W.W, Bountra, C, Bezerra, G.A. | | Deposit date: | 2022-01-20 | | Release date: | 2022-02-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.839 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5SDM
 
 | | PanDDA analysis group deposition -- Crystal Structure of Porphyromonas gingivalis in complex with Z1328078283 | | Descriptor: | Asp/Glu-specific dipeptidyl-peptidase, CHLORIDE ION, N-(cyclopropylmethyl)-2,2,3,3-tetramethylazetidine-1-carboxamide | | Authors: | Tham, C.T, Coker, J.A, Krojer, T, Foster, W.R, Koekemoer, L, Douangamath, A, Talon, R, Fearon, D, von Delft, F, Yue, W.W, Bountra, C, Bezerra, G.A. | | Deposit date: | 2022-01-20 | | Release date: | 2022-02-09 | | Method: | X-RAY DIFFRACTION (2.039 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5SDS
 
 | | PanDDA analysis group deposition of ground-state model of Porphyromonas gingivalis DPP11 | | Descriptor: | Asp/Glu-specific dipeptidyl-peptidase, CHLORIDE ION | | Authors: | Tham, C.T, Coker, J.A, Krojer, T, Foster, W.R, Koekemoer, L, Douangamath, A, Talon, R, Fearon, D, von Delft, F, Yue, W.W, Bountra, C, Bezerra, G.A. | | Deposit date: | 2022-01-20 | | Release date: | 2022-02-09 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | PanDDA analysis group deposition of ground-state model
To Be Published
|
|
5R4V
 
 | | XChem fragment screen -- CRYSTAL STRUCTURE OF THE BROMODOMAIN OF THE HUMAN ATAD2 in complex with N13475a | | Descriptor: | 1,2-ETHANEDIOL, ATPase family AAA domain-containing protein 2, SULFATE ION, ... | | Authors: | Talon, R, Krojer, T, Fairhead, M, Sethi, R, Bradley, A.R, Aimon, A, Collins, P, Brandao-Neto, J, Douangamath, A, Wright, N, MacLean, E, Zhang, R, Dias, A, Brennan, P.E, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2020-02-28 | | Release date: | 2020-05-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | XChem fragment screen
To Be Published
|
|
5R5B
 
 | | PanDDA analysis group deposition -- Crystal Structure of human NUDT22 in complex with N13725a | | Descriptor: | 4-[(phenylmethyl)amino]benzoic acid, DIMETHYL SULFOXIDE, Uridine diphosphate glucose pyrophosphatase NUDT22 | | Authors: | Diaz-Saez, L, Talon, R, Krojer, T, Burgess-Brown, N.A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F, Huber, K.V.M. | | Deposit date: | 2020-02-28 | | Release date: | 2020-07-01 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5RGZ
 
 | | PanDDA analysis group deposition SARS-CoV-2 main protease fragment screen -- Crystal Structure of SARS-CoV-2 main protease in complex with Z1343543528 (Mpro-x2600) | | Descriptor: | 2-(3-cyanophenyl)-N-(pyridin-3-yl)acetamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-05-15 | | Release date: | 2020-05-27 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | PanDDA analysis group deposition SARS-CoV-2 main protease fragment screen
To Be Published
|
|
5RKO
 
 | | PanDDA analysis group deposition -- Crystal Structure of PHIP in complex with Z30620520 | | Descriptor: | PH-interacting protein, cyclopropyl-[4-(4-fluorophenyl)piperazin-1-yl]methanone | | Authors: | Grosjean, H, Aimon, A, Krojer, T, Talon, R, Douangamath, A, Koekemoer, L, Arrowsmith, C.H, Edwards, A, Bountra, C, von Delft, F, Biggin, P.C. | | Deposit date: | 2020-06-02 | | Release date: | 2020-06-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | PanDDA analysis group deposition of ground-state model
To Be Published
|
|
5RKY
 
 | | PanDDA analysis group deposition -- Crystal Structure of PHIP in complex with Z1796014543 | | Descriptor: | 1-[(2-fluorophenyl)methyl]-N-methylcyclopropane-1-carboxamide, PH-interacting protein | | Authors: | Grosjean, H, Aimon, A, Krojer, T, Talon, R, Douangamath, A, Koekemoer, L, Arrowsmith, C.H, Edwards, A, Bountra, C, von Delft, F, Biggin, P.C. | | Deposit date: | 2020-06-02 | | Release date: | 2020-06-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | PanDDA analysis group deposition of ground-state model
To Be Published
|
|
5RLE
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 helicase in complex with Z1429867185 | | Descriptor: | 4-methoxy-1H-indole, Helicase, PHOSPHATE ION, ... | | Authors: | Newman, J.A, Yosaatmadja, Y, Douangamath, A, Aimon, A, Powell, A.J, Dias, A, Fearon, D, Dunnett, L, Brandao-Neto, J, Krojer, T, Skyner, R, Gorrie-Stone, T, Thompson, W, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2020-09-16 | | Release date: | 2020-09-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.268 Å) | | Cite: | Structure, mechanism and crystallographic fragment screening of the SARS-CoV-2 NSP13 helicase.
Nat Commun, 12, 2021
|
|
5RMC
 
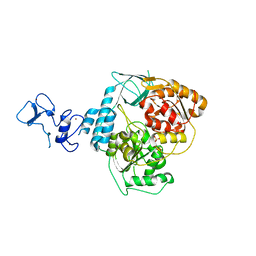 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 helicase in complex with Z24758179 | | Descriptor: | Helicase, PHOSPHATE ION, ZINC ION, ... | | Authors: | Newman, J.A, Yosaatmadja, Y, Douangamath, A, Aimon, A, Powell, A.J, Dias, A, Fearon, D, Dunnett, L, Brandao-Neto, J, Krojer, T, Skyner, R, Gorrie-Stone, T, Thompson, W, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2020-09-16 | | Release date: | 2020-09-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure, mechanism and crystallographic fragment screening of the SARS-CoV-2 NSP13 helicase.
Nat Commun, 12, 2021
|
|
5RH2
 
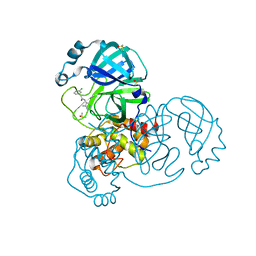 | | PanDDA analysis group deposition SARS-CoV-2 main protease fragment screen -- Crystal Structure of SARS-CoV-2 main protease in complex with Z1129289650 (Mpro-x2646) | | Descriptor: | 2-(3-chlorophenyl)-N-(4-methylpyridin-3-yl)acetamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-05-15 | | Release date: | 2020-05-27 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.827 Å) | | Cite: | PanDDA analysis group deposition SARS-CoV-2 main protease fragment screen
To Be Published
|
|
5RYS
 
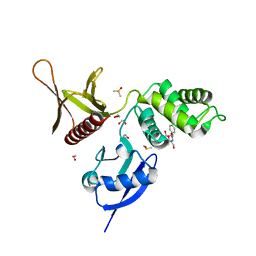 | | EPB41L3 PanDDA analysis group deposition -- Crystal Structure of the FERM domain of human EPB41L3 in complex with Z1593306637 | | Descriptor: | 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, Isoform 2 of Band 4.1-like protein 3, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Newman, J.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gileadi, O. | | Deposit date: | 2020-10-30 | | Release date: | 2020-11-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | EPB41L3 PanDDA analysis group deposition
To Be Published
|
|
5RZ4
 
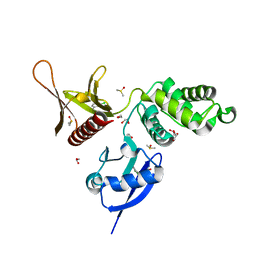 | | EPB41L3 PanDDA analysis group deposition -- Crystal Structure of the FERM domain of human EPB41L3 in complex with Z2856434909 | | Descriptor: | (2R)-1-{[(2,6-dichlorophenyl)methyl]amino}propan-2-ol, 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Newman, J.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gileadi, O. | | Deposit date: | 2020-10-30 | | Release date: | 2020-11-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | EPB41L3 PanDDA analysis group deposition
To Be Published
|
|
5RZL
 
 | | EPB41L3 PanDDA analysis group deposition -- Crystal Structure of the FERM domain of human EPB41L3 in complex with Z1492796719 | | Descriptor: | 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, Isoform 2 of Band 4.1-like protein 3, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Newman, J.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gileadi, O. | | Deposit date: | 2020-10-30 | | Release date: | 2020-11-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | EPB41L3 PanDDA analysis group deposition
To Be Published
|
|
5RM5
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 helicase in complex with Z373768900 | | Descriptor: | Helicase, N-(1-ethyl-1H-pyrazol-4-yl)cyclobutanecarboxamide, PHOSPHATE ION, ... | | Authors: | Newman, J.A, Yosaatmadja, Y, Douangamath, A, Aimon, A, Powell, A.J, Dias, A, Fearon, D, Dunnett, L, Brandao-Neto, J, Krojer, T, Skyner, R, Gorrie-Stone, T, Thompson, W, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2020-09-16 | | Release date: | 2020-09-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structure, mechanism and crystallographic fragment screening of the SARS-CoV-2 NSP13 helicase.
Nat Commun, 12, 2021
|
|
5S18
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with EN300-321461 | | Descriptor: | 6,7-dihydro-5H-pyrrolo[2,3-d]pyrimidine, DIMETHYL SULFOXIDE, Non-structural protein 3 | | Authors: | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | Deposit date: | 2020-11-02 | | Release date: | 2021-01-13 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5RMI
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 helicase in complex with Z53860899 | | Descriptor: | Helicase, PHOSPHATE ION, ZINC ION, ... | | Authors: | Newman, J.A, Yosaatmadja, Y, Douangamath, A, Aimon, A, Powell, A.J, Dias, A, Fearon, D, Dunnett, L, Brandao-Neto, J, Krojer, T, Skyner, R, Gorrie-Stone, T, Thompson, W, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2020-09-16 | | Release date: | 2020-09-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.116 Å) | | Cite: | Structure, mechanism and crystallographic fragment screening of the SARS-CoV-2 NSP13 helicase.
Nat Commun, 12, 2021
|
|
5R50
 
 | | PanDDA analysis group deposition -- Crystal Structure of human NUDT22 in complex with N13371a | | Descriptor: | DIMETHYL SULFOXIDE, N-[4-(aminomethyl)phenyl]methanesulfonamide, Uridine diphosphate glucose pyrophosphatase NUDT22 | | Authors: | Diaz-Saez, L, Talon, R, Krojer, T, Burgess-Brown, N.A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F, Huber, K.V.M. | | Deposit date: | 2020-02-28 | | Release date: | 2020-07-01 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5R5I
 
 | | PanDDA analysis group deposition -- Crystal Structure of human NUDT22 in complex with N06325b | | Descriptor: | 1-ethanoylpiperidine-4-carbonitrile, DIMETHYL SULFOXIDE, Uridine diphosphate glucose pyrophosphatase NUDT22 | | Authors: | Diaz-Saez, L, Talon, R, Krojer, T, Burgess-Brown, N.A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F, Huber, K.V.M. | | Deposit date: | 2020-02-28 | | Release date: | 2020-07-01 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5R5W
 
 | | PanDDA analysis group deposition -- Crystal Structure of FIBRINOGEN-LIKE GLOBE DOMAIN OF HUMAN TENASCIN-C in complex with Z2856434942 | | Descriptor: | Tenascin C (Hexabrachion), isoform CRA_a, ~{N}-(4-fluorophenyl)-2-pyrrolidin-1-yl-ethanamide | | Authors: | Coker, J.A, Bezerra, G.A, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Yue, W.W, Marsden, B.D. | | Deposit date: | 2020-02-28 | | Release date: | 2020-10-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5RVY
 
 | | PanDDA analysis group deposition -- Crystal Structure of DHTKD1 in complex with Z437516460 | | Descriptor: | MAGNESIUM ION, N-[(6-methylpyridin-3-yl)methyl]cyclobutanecarboxamide, Probable 2-oxoglutarate dehydrogenase E1 component DHKTD1, ... | | Authors: | Bezerra, G.A, Foster, W.R, Bailey, H.J, Shrestha, L, Krojer, T, Brandao-Neto, J, Douangamath, A, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Yue, W.W. | | Deposit date: | 2020-10-27 | | Release date: | 2020-11-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.609 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
