5YF6
 
 | |
5YF7
 
 | |
5YF5
 
 | |
8HZT
 
 | |
4MSV
 
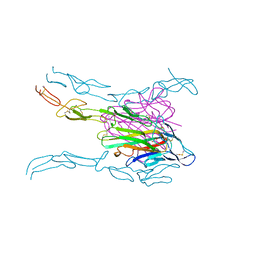 | | Crystal structure of FASL and DcR3 complex | | 分子名称: | GLYCEROL, MAGNESIUM ION, Tumor necrosis factor ligand superfamily member 6, ... | | 著者 | Liu, W, Ramagopal, U.A, Zhan, C, Bonanno, J.B, Bhosle, R.C, Nathenson, S.G, Almo, S.C, Atoms-to-Animals: The Immune Function Network (IFN), New York Structural Genomics Research Consortium (NYSGRC) | | 登録日 | 2013-09-18 | | 公開日 | 2013-11-27 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal Structure of the Complex of Human FasL and Its Decoy Receptor DcR3.
Structure, 24, 2016
|
|
6AE4
 
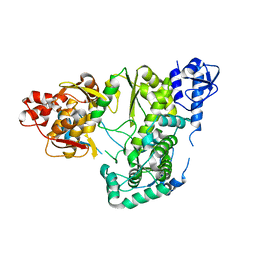 | |
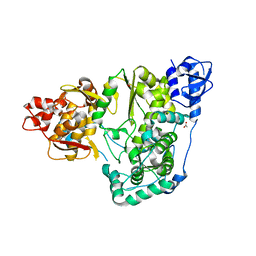
6AE6
 
 | |
6AE7
 
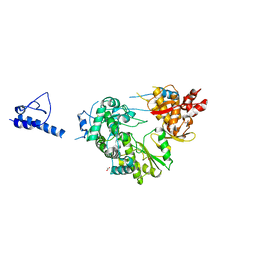 | |
6AE5
 
 | |
8I0A
 
 | |
6JJ0
 
 | |
7W12
 
 | | Complex structure of alginate lyase AlyB-OU02 with G9 | | 分子名称: | Alginate lyase, SULFATE ION, alpha-L-gulopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid | | 著者 | Liu, W.Z, Lyu, Q.Q, Zhang, K.K. | | 登録日 | 2021-11-19 | | 公開日 | 2022-11-16 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Determination of oligosaccharide product distributions of PL7 alginate lyases by their structural elements.
Commun Biol, 5, 2022
|
|
7W16
 
 | | Complex structure of alginate lyase AlyV with M8 | | 分子名称: | GLYCEROL, alginate lyase, beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid | | 著者 | Liu, W.Z, Lyu, Q.Q, Li, Z.J, Zhang, K.K. | | 登録日 | 2021-11-19 | | 公開日 | 2022-08-17 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Determination of oligosaccharide product distributions of PL7 alginate lyases by their structural elements.
Commun Biol, 5, 2022
|
|
7W13
 
 | | Complex structure of alginate lyase PyAly with M8 | | 分子名称: | Alginate lyase, beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid | | 著者 | Liu, W.Z, Lyu, Q.Q, Li, Z.J, Zhang, K.K. | | 登録日 | 2021-11-19 | | 公開日 | 2022-08-17 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Determination of oligosaccharide product distributions of PL7 alginate lyases by their structural elements.
Commun Biol, 5, 2022
|
|
8HZQ
 
 | |
6NG9
 
 | |
6NGG
 
 | |
7W18
 
 | | Complex structure of alginate lyase PyAly with M5 | | 分子名称: | Alginate lyase, beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid | | 著者 | Liu, W.Z, Lyu, Q.Q, Zhang, K.K. | | 登録日 | 2021-11-19 | | 公開日 | 2022-08-17 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Determination of oligosaccharide product distributions of PL7 alginate lyases by their structural elements.
Commun Biol, 5, 2022
|
|
6NG3
 
 | | Crystal structure of human CD160 and HVEM complex | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CD160 antigen,Tumor necrosis factor receptor superfamily member 14, MAGNESIUM ION, ... | | 著者 | Liu, W, Bonanno, J, Almo, S.C. | | 登録日 | 2018-12-21 | | 公開日 | 2019-07-03 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.88 Å) | | 主引用文献 | Structural Basis of CD160:HVEM Recognition.
Structure, 27, 2019
|
|
7EKJ
 
 | |
7LW2
 
 | |
4H37
 
 | | Crystal structure of a voltage-gated K+ channel pore domain in a closed state in lipid membranes | | 分子名称: | Lmo2059 protein, POTASSIUM ION | | 著者 | Santos, J.S, Asmar-Rovira, G.A, Han, G.W, Liu, W, Syeda, R, Cherezov, V, Baker, K.A, Stevens, R.C, Montal, M. | | 登録日 | 2012-09-13 | | 公開日 | 2012-11-07 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (3.35 Å) | | 主引用文献 | Crystal Structure of a Voltage-gated K+ Channel Pore Module in a Closed State in Lipid Membranes.
J.Biol.Chem., 287, 2012
|
|
6O2L
 
 | |
4UXL
 
 | | Structure of Human ROS1 Kinase Domain in Complex with PF-06463922 | | 分子名称: | (10R)-7-amino-12-fluoro-2,10,16-trimethyl-15-oxo-10,15,16,17-tetrahydro-2H-8,4-(metheno)pyrazolo[4,3-h][2,5,11]benzoxadiazacyclotetradecine-3-carbonitrile, PROTO-ONCOGENE TYROSINE-PROTEIN KINASE ROS | | 著者 | McTigue, M, Deng, Y, Liu, W, Brooun, A, Stewart, A. | | 登録日 | 2014-08-25 | | 公開日 | 2015-03-11 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Pf-06463922 is a Potent and Selective Next-Generation Ros1/Alk Inhibitor Capable of Blocking Crizotinib-Resistant Ros1 Mutations.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5DTH
 
 | |
