5ZRY
 
 | | Crystal Structure of EphA6/Odin Complex | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Ankyrin repeat and SAM domain-containing protein 1A,Ephrin type-A receptor 6, ... | | Authors: | Wang, Y, Shang, Y, Li, J, Chen, W, Li, G, Wan, J, Liu, W, Zhang, M. | | Deposit date: | 2018-04-25 | | Release date: | 2018-05-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Specific Eph receptor-cytoplasmic effector signaling mediated by SAM-SAM domain interactions.
Elife, 7, 2018
|
|
5XK6
 
 | | Structure of a prenyltransferase soaked with IPP | | Descriptor: | MAGNESIUM ION, PYROPHOSPHATE 2-, SULFATE ION, ... | | Authors: | Ko, T.P, Guo, R.T, Liu, W, Chen, C.C, Gao, J. | | Deposit date: | 2017-05-05 | | Release date: | 2018-01-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | "Head-to-Middle" and "Head-to-Tail" cis-Prenyl Transferases: Structure of Isosesquilavandulyl Diphosphate Synthase.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
5XK3
 
 | | Crystal structure of apo form Isosesquilavandulyl Diphosphate Synthase from Streptomyces sp. strain CNH-189 | | Descriptor: | SULFATE ION, Undecaprenyl diphosphate synthase | | Authors: | Ko, T.P, Guo, R.T, Liu, W, Chen, C.C, Gao, J. | | Deposit date: | 2017-05-05 | | Release date: | 2018-01-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.996 Å) | | Cite: | "Head-to-Middle" and "Head-to-Tail" cis-Prenyl Transferases: Structure of Isosesquilavandulyl Diphosphate Synthase.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
6IME
 
 | | Rv2361c complex with substrate analogues | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 3-methylbut-3-enylsulfanyl(phosphonooxy)phosphinic acid, CARBONATE ION, ... | | Authors: | Ko, T.-P, Guo, R.-T, Chen, C.-C, Liu, W. | | Deposit date: | 2018-10-22 | | Release date: | 2019-09-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Substrate-analogue complex structure of Mycobacterium tuberculosis decaprenyl diphosphate synthase.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
7XDR
 
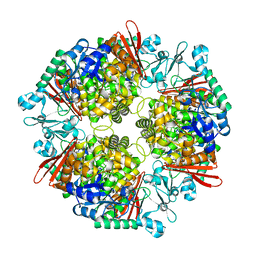 | | Crystal structure of a glucosylglycerol phosphorylase from Marinobacter adhaerens | | Descriptor: | Glucosylglycerol phosphorylase | | Authors: | Wei, H.L, Li, Q, Yang, J.G, Liu, W.D, Sun, Y.X. | | Deposit date: | 2022-03-28 | | Release date: | 2023-02-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Protein Engineering of Glucosylglycerol Phosphorylase Facilitating Efficient and Highly Regio- and Stereoselective Glycosylation of Polyols in a Synthetic System.
Acs Catalysis, 2022
|
|
7W66
 
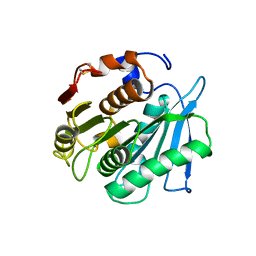 | | Crystal structure of a PSH1 mutant in complex with ligand | | Descriptor: | PSH1, bis(2-hydroxyethyl) benzene-1,4-dicarboxylate | | Authors: | Gao, J, Lara, P, Li, Z.S, Han, X, Wei, R, Liu, W.D. | | Deposit date: | 2021-12-01 | | Release date: | 2022-09-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Multiple Substrate Binding Mode-Guided Engineering of a Thermophilic PET Hydrolase.
Acs Catalysis, 12, 2022
|
|
7W6C
 
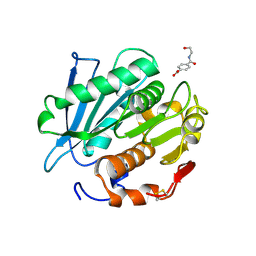 | | Crystal structure of a PSH1 in complex with ligand J1K | | Descriptor: | 4-(2-hydroxyethylcarbamoyl)benzoic acid, PSH1 | | Authors: | Gao, J, Lara, P, Li, Z.S, Han, X, Wei, R, Liu, W.D. | | Deposit date: | 2021-12-01 | | Release date: | 2022-09-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Multiple Substrate Binding Mode-Guided Engineering of a Thermophilic PET Hydrolase.
Acs Catalysis, 12, 2022
|
|
7W69
 
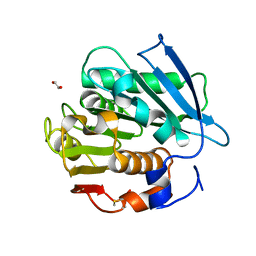 | | Crystal structure of a PSH1 mutant in complex with EDO | | Descriptor: | 1,2-ETHANEDIOL, PSH1 | | Authors: | Gao, J, Lara, P, Li, Z.S, Han, X, Wei, R, Liu, W.D. | | Deposit date: | 2021-12-01 | | Release date: | 2022-09-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Multiple Substrate Binding Mode-Guided Engineering of a Thermophilic PET Hydrolase.
Acs Catalysis, 12, 2022
|
|
7W6O
 
 | | Crystal structure of a PSH1 in complex with J1K | | Descriptor: | 4-(2-hydroxyethylcarbamoyl)benzoic acid, PSH1 | | Authors: | Gao, J, Lara, P, Li, Z.S, Han, X, Wei, R, Liu, W.D. | | Deposit date: | 2021-12-02 | | Release date: | 2022-09-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Multiple Substrate Binding Mode-Guided Engineering of a Thermophilic PET Hydrolase.
Acs Catalysis, 12, 2022
|
|
7W6Q
 
 | | Crystal structure of a PSH1 in complex with ligand J1K | | Descriptor: | 4-(2-hydroxyethylcarbamoyl)benzoic acid, PSH1 | | Authors: | Gao, J, Lara, P, Li, Z.S, Han, X, Wei, R, Liu, W.D. | | Deposit date: | 2021-12-02 | | Release date: | 2022-09-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Multiple Substrate Binding Mode-Guided Engineering of a Thermophilic PET Hydrolase.
Acs Catalysis, 12, 2022
|
|
7EMF
 
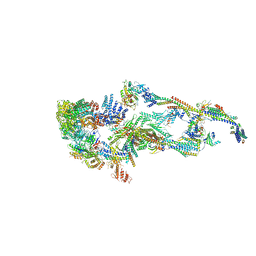 | | Human Mediator (deletion of MED1-IDR) in a Tail-extended conformation | | Descriptor: | Isoform 2 of Mediator of RNA polymerase II transcription subunit 16, Isoform 2 of Mediator of RNA polymerase II transcription subunit 8, Mediator of RNA polymerase II transcription subunit 1, ... | | Authors: | Yin, X, Li, J, Wu, Z, Liu, W, Xu, Y. | | Deposit date: | 2021-04-13 | | Release date: | 2021-05-05 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structures of the human Mediator and Mediator-bound preinitiation complex.
Science, 372, 2021
|
|
7ENJ
 
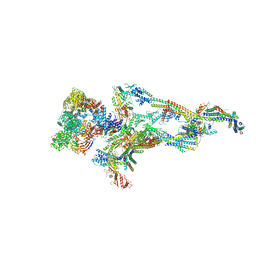 | | Human Mediator (deletion of MED1-IDR) in a Tail-bent conformation (MED-B) | | Descriptor: | Isoform 2 of Mediator of RNA polymerase II transcription subunit 8, Mediator of RNA polymerase II transcription subunit 1, Mediator of RNA polymerase II transcription subunit 10, ... | | Authors: | Yin, X, Li, J, Wu, Z, Liu, W, Xu, Y. | | Deposit date: | 2021-04-17 | | Release date: | 2021-05-19 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structures of the human Mediator and Mediator-bound preinitiation complex.
Science, 372, 2021
|
|
7ENA
 
 | | TFIID-based PIC-Mediator holo-complex in pre-assembled state (pre-hPIC-MED) | | Descriptor: | CDK-activating kinase assembly factor MAT1, Cyclin-H, Cyclin-dependent kinase 7, ... | | Authors: | Chen, X, Qi, Y, Wang, X, Wu, Z, Yin, X, Li, J, Liu, W, Xu, Y. | | Deposit date: | 2021-04-16 | | Release date: | 2021-05-26 | | Last modified: | 2021-06-16 | | Method: | ELECTRON MICROSCOPY (4.07 Å) | | Cite: | Structures of the human Mediator and Mediator-bound preinitiation complex.
Science, 372, 2021
|
|
7ENC
 
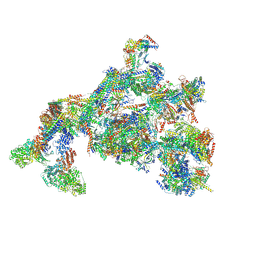 | | TFIID-based PIC-Mediator holo-complex in fully-assembled state (hPIC-MED) | | Descriptor: | CDK-activating kinase assembly factor MAT1, Cyclin-H, Cyclin-dependent kinase 7, ... | | Authors: | Chen, X, Qi, Y, Wang, X, Wu, Z, Yin, X, Li, J, Liu, W, Xu, Y. | | Deposit date: | 2021-04-16 | | Release date: | 2021-05-26 | | Last modified: | 2021-06-16 | | Method: | ELECTRON MICROSCOPY (4.13 Å) | | Cite: | Structures of the human Mediator and Mediator-bound preinitiation complex.
Science, 372, 2021
|
|
7Y8M
 
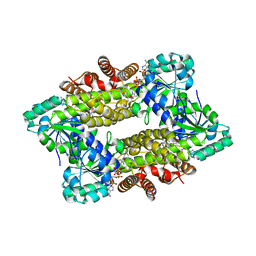 | | Structure of ScIRED-R2-V3 from Streptomyces clavuligerus in complex with 5-(3-fluorophenyl)-3,4-dihydro-2H-pyrrole | | Descriptor: | 2-[2,5-bis(fluoranyl)phenyl]pyrrolidine, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, reductase | | Authors: | Zhang, L.L, Liu, W.D, Shi, M, Huang, J.W, Yang, Y, Chen, C.C, Guo, R.T. | | Deposit date: | 2022-06-24 | | Release date: | 2023-06-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Engineered Imine Reductase for Larotrectinib Intermediate Manufacture
Acs Catalysis, 12, 2022
|
|
5XK8
 
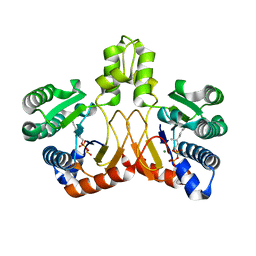 | | Crystal structure of Isosesquilavandulyl Diphosphate Synthase from Streptomyces sp. strain CNH-189 in complex with GPP | | Descriptor: | GERANYL DIPHOSPHATE, MAGNESIUM ION, Undecaprenyl diphosphate synthase | | Authors: | Ko, T.P, Guo, R.T, Liu, W, Chen, C.C, Gao, J. | | Deposit date: | 2017-05-05 | | Release date: | 2018-01-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.304 Å) | | Cite: | "Head-to-Middle" and "Head-to-Tail" cis-Prenyl Transferases: Structure of Isosesquilavandulyl Diphosphate Synthase.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
7Y8O
 
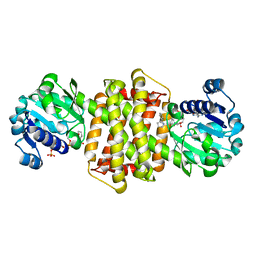 | | Structure of ScIRED-R3-V4 from Streptomyces clavuligerus in complex with 5-(3-fluorophenyl)-3,4-dihydro-2H-pyrrole | | Descriptor: | 2-[2,5-bis(fluoranyl)phenyl]pyrrolidine, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SciR | | Authors: | Zhang, L.L, Liu, W.D, Shi, M, Huang, J.W, Yang, Y, Chen, C.C, Guo, R.T. | | Deposit date: | 2022-06-24 | | Release date: | 2023-03-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of ScIRED-R3-V4 from Streptomyces clavuligerus in complex with 5-(3-fluorophenyl)-3,4-dihydro-2H-pyrrole
to be published
|
|
8H8Y
 
 | | Crystal structure of AbHheG from Acidimicrobiia bacterium | | Descriptor: | GLYCEROL, alpha/beta hydrolase | | Authors: | Zhou, C.H, Chen, X, Han, X, Liu, W.D, Wu, Q.Q, Zhu, D.M, Ma, Y.H. | | Deposit date: | 2022-10-24 | | Release date: | 2023-08-02 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Flipping the Substrate Creates a Highly Selective Halohydrin Dehalogenase for the Synthesis of Chiral 4-Aryl-2-oxazolidinones from Readily Available Epoxides
Acs Catalysis, 13, 2023
|
|
8HQP
 
 | | Crystal structure of AbHheG mutant from Acidimicrobiia bacterium | | Descriptor: | AbHheG_m | | Authors: | Zhou, C.H, Chen, X, Han, X, Liu, W.D, Wu, Q.Q, Zhu, D.M, Ma, Y.H. | | Deposit date: | 2022-12-13 | | Release date: | 2023-08-02 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Flipping the Substrate Creates a Highly Selective Halohydrin Dehalogenase for the Synthesis of Chiral 4-Aryl-2-oxazolidinones from Readily Available Epoxides
Acs Catalysis, 13, 2023
|
|
7DCJ
 
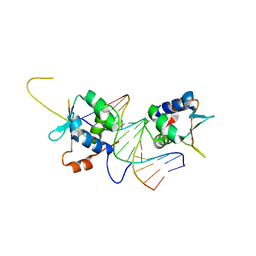 | |
7DCT
 
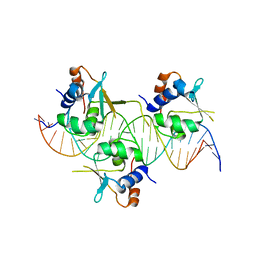 | | Crystal structure of HSF1 DNA-binding domain in complex with 3-site HSE DNA (24 bp) | | Descriptor: | DNA (5'-D(*AP*CP*TP*CP*GP*CP*GP*AP*AP*TP*AP*TP*TP*CP*TP*AP*GP*AP*AP*CP*GP*CP*AP*C)-3'), DNA (5'-D(*TP*GP*TP*GP*CP*GP*TP*TP*CP*TP*AP*GP*AP*AP*TP*AP*TP*TP*CP*GP*CP*GP*AP*G)-3'), Heat shock factor protein 1, ... | | Authors: | Feng, N, Liu, W. | | Deposit date: | 2020-10-27 | | Release date: | 2021-07-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Structures of heat shock factor trimers bound to DNA.
Iscience, 24, 2021
|
|
7DCU
 
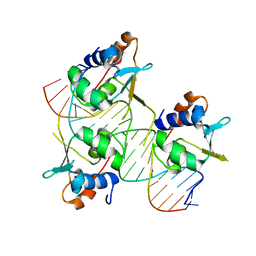 | | Crystal structure of HSF2 DNA-binding domain in complex with 3-site HSE DNA (21 bp) | | Descriptor: | DNA (5'-D(*AP*CP*CP*GP*CP*GP*AP*AP*TP*AP*TP*TP*CP*TP*AP*GP*AP*AP*CP*GP*C)-3'), DNA (5'-D(*TP*GP*CP*GP*TP*TP*CP*TP*AP*GP*AP*AP*TP*AP*TP*TP*CP*GP*CP*GP*G)-3'), Heat shock factor protein 2, ... | | Authors: | Feng, N, Liu, W. | | Deposit date: | 2020-10-27 | | Release date: | 2021-07-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structures of heat shock factor trimers bound to DNA.
Iscience, 24, 2021
|
|
7DTL
 
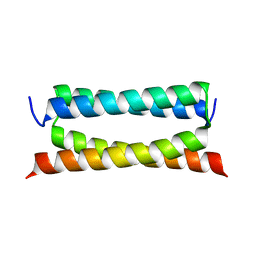 | | Crystal structure of PSK, an antimicrobial peptide from Chrysomya megacephala | | Descriptor: | PSK | | Authors: | Xiao, C, Xiao, Z, Wang, S, Liu, W. | | Deposit date: | 2021-01-05 | | Release date: | 2021-07-14 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal and solution structures of a novel antimicrobial peptide from Chrysomya megacephala.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
8H83
 
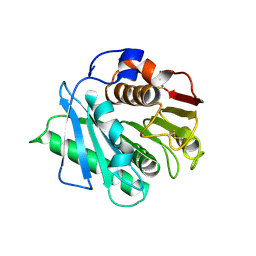 | | Crystal structure of a IsPETase variant V22 from Ideonella sakaiensis | | Descriptor: | Poly(ethylene terephthalate) hydrolase | | Authors: | Wei, H.L, Gao, S.F, Li, Q, Han, X, Gao, J, Liu, W.D. | | Deposit date: | 2022-10-21 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal structure of a IsPETase variant V22 from Ideonella sakaiensis
to be published
|
|
8IJ7
 
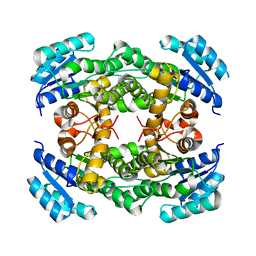 | |
