5XH2
 
 | | Crystal structure of a novel PET hydrolase R103G/S131A mutant in complex with pNP from Ideonella sakaiensis 201-F6 | | Descriptor: | P-NITROPHENOL, Poly(ethylene terephthalate) hydrolase, SULFATE ION | | Authors: | Han, X, Liu, W.D, Zheng, Y.Y, Chen, C.C, Guo, R.T. | | Deposit date: | 2017-04-19 | | Release date: | 2017-12-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural insight into catalytic mechanism of PET hydrolase
Nat Commun, 8, 2017
|
|
1AJ4
 
 | | STRUCTURE OF CALCIUM-SATURATED CARDIAC TROPONIN C, NMR, 1 STRUCTURE | | Descriptor: | CALCIUM ION, TROPONIN C | | Authors: | Sia, S.K, Li, M.X, Spyracopoulos, L, Gagne, S.M, Liu, W, Putkey, J.A, Sykes, B.D. | | Deposit date: | 1997-05-14 | | Release date: | 1998-05-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of cardiac muscle troponin C unexpectedly reveals a closed regulatory domain.
J.Biol.Chem., 272, 1997
|
|
5XH3
 
 | | Crystal structure of a novel PET hydrolase R103G/S131A mutant in complex with HEMT from Ideonella sakaiensis 201-F6 | | Descriptor: | GLYCEROL, O 4-(2-hydroxyethyl) O 1-methyl benzene-1,4-dicarboxylate, Poly(ethylene terephthalate) hydrolase, ... | | Authors: | Han, X, Liu, W.D, Zheng, Y.Y, Chen, C.C, Guo, R.T. | | Deposit date: | 2017-04-19 | | Release date: | 2017-12-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural insight into catalytic mechanism of PET hydrolase
Nat Commun, 8, 2017
|
|
7RSM
 
 | |
6IHI
 
 | | Crystal structure of RasADH 3B3/I91V from Ralstonia.sp in complex with NADPH and A6O | | Descriptor: | (2R,3S)-2-ethyl-2-[(2E)-2-(6-methoxy-3,4-dihydro-2H-naphthalen-1-ylidene)ethyl]-3-oxidanyl-cyclopentan-1-one, Alclohol dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zhang, H.L, Chen, X, Liu, W.D, Wu, Q.Q, Zhu, D.M. | | Deposit date: | 2018-09-30 | | Release date: | 2019-10-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Efficient reductive desymmetrization of bulky 1,3-cyclodiketones enabled by structure-guided directed evolution of a carbonyl reductase.
Nat Catal, 2, 2019
|
|
6IHH
 
 | | Crystal structure of RasADH F12 from Ralstonia.sp in complex with NADPH and A6O | | Descriptor: | (2R,3S)-2-ethyl-2-[(2E)-2-(6-methoxy-3,4-dihydro-2H-naphthalen-1-ylidene)ethyl]-3-oxidanyl-cyclopentan-1-one, Alclohol dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zhang, H.L, Chen, X, Liu, W.D, Wu, Q.Q, Zhu, D.M. | | Deposit date: | 2018-09-30 | | Release date: | 2019-10-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Efficient reductive desymmetrization of bulky 1,3-cyclodiketones enabled by structure-guided directed evolution of a carbonyl reductase.
Nat Catal, 2, 2019
|
|
7RVO
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI13 | | Descriptor: | 3C-like proteinase, N-[(benzyloxy)carbonyl]-L-valyl-3-cyclopropyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-alaninamide | | Authors: | Yang, K, Sankaran, B, Liu, W. | | Deposit date: | 2021-08-19 | | Release date: | 2022-07-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A multi-pronged evaluation of aldehyde-based tripeptidyl main protease inhibitors as SARS-CoV-2 antivirals.
Eur.J.Med.Chem., 240, 2022
|
|
7RVS
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI19 | | Descriptor: | 3C-like proteinase, N-[(benzyloxy)carbonyl]-3-methyl-L-valyl-3-cyclopropyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-alaninamide | | Authors: | Yang, K, Liu, W. | | Deposit date: | 2021-08-19 | | Release date: | 2022-07-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A multi-pronged evaluation of aldehyde-based tripeptidyl main protease inhibitors as SARS-CoV-2 antivirals.
Eur.J.Med.Chem., 240, 2022
|
|
7RVR
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI18 | | Descriptor: | 3C-like proteinase, N-[(benzyloxy)carbonyl]-3-methyl-L-valyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-4-methyl-L-leucinamide | | Authors: | Yang, K, Liu, W. | | Deposit date: | 2021-08-19 | | Release date: | 2022-07-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | A multi-pronged evaluation of aldehyde-based tripeptidyl main protease inhibitors as SARS-CoV-2 antivirals.
Eur.J.Med.Chem., 240, 2022
|
|
7RW0
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI27 | | Descriptor: | 3C-like proteinase, N-{[(3-chlorophenyl)methoxy]carbonyl}-L-valyl-3-cyclohexyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-alaninamide | | Authors: | Yang, K, Liu, W. | | Deposit date: | 2021-08-19 | | Release date: | 2022-07-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A multi-pronged evaluation of aldehyde-based tripeptidyl main protease inhibitors as SARS-CoV-2 antivirals.
Eur.J.Med.Chem., 240, 2022
|
|
7RVM
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI11 | | Descriptor: | 3C-like proteinase, N-[(benzyloxy)carbonyl]-L-valyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-4-methyl-L-leucinamide | | Authors: | Yang, K, Liu, W. | | Deposit date: | 2021-08-19 | | Release date: | 2022-07-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A multi-pronged evaluation of aldehyde-based tripeptidyl main protease inhibitors as SARS-CoV-2 antivirals.
Eur.J.Med.Chem., 240, 2022
|
|
7RVW
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI23 | | Descriptor: | 3C-like proteinase, benzyl (1-{[(2S)-3-cyclohexyl-1-({(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}amino)-1-oxopropan-2-yl]carbamoyl}cyclopropyl)carbamate | | Authors: | Yang, K, Liu, W. | | Deposit date: | 2021-08-19 | | Release date: | 2022-07-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A multi-pronged evaluation of aldehyde-based tripeptidyl main protease inhibitors as SARS-CoV-2 antivirals.
Eur.J.Med.Chem., 240, 2022
|
|
7RVU
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI21 | | Descriptor: | 3C-like proteinase, N-[(benzyloxy)carbonyl]-3-methyl-L-isovalyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-4-methyl-L-leucinamide | | Authors: | Yang, K, Sankaran, B, Liu, W. | | Deposit date: | 2021-08-19 | | Release date: | 2022-07-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A multi-pronged evaluation of aldehyde-based tripeptidyl main protease inhibitors as SARS-CoV-2 antivirals.
Eur.J.Med.Chem., 240, 2022
|
|
7RW1
 
 | |
7RVZ
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI26 | | Descriptor: | 3C-like proteinase, O-tert-butyl-N-{[(3-chlorophenyl)methoxy]carbonyl}-L-threonyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide | | Authors: | Yang, K, Sankaran, B, Liu, W. | | Deposit date: | 2021-08-19 | | Release date: | 2022-07-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A multi-pronged evaluation of aldehyde-based tripeptidyl main protease inhibitors as SARS-CoV-2 antivirals.
Eur.J.Med.Chem., 240, 2022
|
|
7RVV
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI22 | | Descriptor: | 3C-like proteinase, N-[(benzyloxy)carbonyl]-2-methyl-L-alanyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-4-methyl-L-leucinamide | | Authors: | Yang, K, Liu, W. | | Deposit date: | 2021-08-19 | | Release date: | 2022-07-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A multi-pronged evaluation of aldehyde-based tripeptidyl main protease inhibitors as SARS-CoV-2 antivirals.
Eur.J.Med.Chem., 240, 2022
|
|
7RVQ
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI16 | | Descriptor: | 3C-like proteinase, N-[(benzyloxy)carbonyl]-O-tert-butyl-L-threonyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-4-methyl-L-leucinamide | | Authors: | Yang, K, Liu, W. | | Deposit date: | 2021-08-19 | | Release date: | 2022-07-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | A multi-pronged evaluation of aldehyde-based tripeptidyl main protease inhibitors as SARS-CoV-2 antivirals.
Eur.J.Med.Chem., 240, 2022
|
|
7RVX
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI24 | | Descriptor: | 3C-like proteinase, benzyl [(1S)-1-cyclopropyl-2-{[(2S)-3-cyclopropyl-1-({(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}amino)-1-oxopropan-2-yl]amino}-2-oxoethyl]carbamate | | Authors: | Yang, K, Liu, W. | | Deposit date: | 2021-08-19 | | Release date: | 2022-07-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A multi-pronged evaluation of aldehyde-based tripeptidyl main protease inhibitors as SARS-CoV-2 antivirals.
Eur.J.Med.Chem., 240, 2022
|
|
7RVP
 
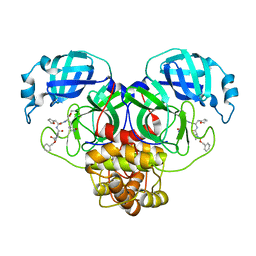 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI14 | | Descriptor: | 3C-like proteinase, N-[(benzyloxy)carbonyl]-L-valyl-3-furan-2-yl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-alaninamide | | Authors: | Yang, K, Sankaran, B, Liu, W. | | Deposit date: | 2021-08-19 | | Release date: | 2022-07-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A multi-pronged evaluation of aldehyde-based tripeptidyl main protease inhibitors as SARS-CoV-2 antivirals.
Eur.J.Med.Chem., 240, 2022
|
|
7RVT
 
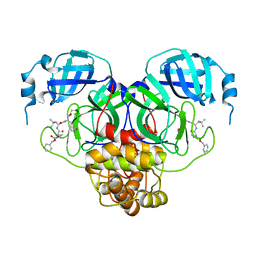 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI20 | | Descriptor: | 3C-like proteinase, N~2~-[(2S)-2-{[(benzyloxy)carbonyl]amino}-2-cyclopropylacetyl]-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-4-methyl-L-leucinamide | | Authors: | Yang, K, Sankaran, B, Liu, W. | | Deposit date: | 2021-08-19 | | Release date: | 2022-07-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A multi-pronged evaluation of aldehyde-based tripeptidyl main protease inhibitors as SARS-CoV-2 antivirals.
Eur.J.Med.Chem., 240, 2022
|
|
7RVN
 
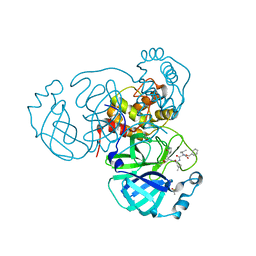 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI12 | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, N-[(benzyloxy)carbonyl]-L-valyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-4-methylidene-L-norvalinamide | | Authors: | Yang, K, Liu, W. | | Deposit date: | 2021-08-19 | | Release date: | 2022-07-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | A multi-pronged evaluation of aldehyde-based tripeptidyl main protease inhibitors as SARS-CoV-2 antivirals.
Eur.J.Med.Chem., 240, 2022
|
|
7RVY
 
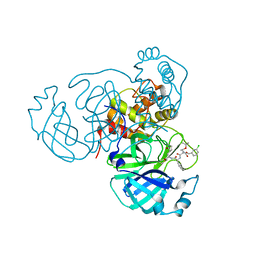 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI25 | | Descriptor: | 3C-like proteinase, O-tert-butyl-N-{[(3-chlorophenyl)methoxy]carbonyl}-L-threonyl-3-cyclohexyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-alaninamide | | Authors: | Yang, K, Liu, W. | | Deposit date: | 2021-08-19 | | Release date: | 2022-07-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A multi-pronged evaluation of aldehyde-based tripeptidyl main protease inhibitors as SARS-CoV-2 antivirals.
Eur.J.Med.Chem., 240, 2022
|
|
5YNW
 
 | | Crystal structure of an aromatic prenyltransferase FAMD1 from Fischerella ambigua UTEX 1903 in complex with DMASPP and INN | | Descriptor: | 3-[(Z)-2-isocyanoethenyl]-1H-indole, DIMETHYLALLYL S-THIOLODIPHOSPHATE, GLYCEROL, ... | | Authors: | Wang, J, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2017-10-25 | | Release date: | 2018-08-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural insight into a novel indole prenyltransferase in hapalindole-type alkaloid biosynthesis.
Biochem. Biophys. Res. Commun., 495, 2018
|
|
2HZ8
 
 | | QM/MM structure refined from NMR-structure of a single chain diiron protein | | Descriptor: | De novo designed diiron protein, ZINC ION | | Authors: | Calhoun, J.R, Liu, W, Spiegel, K, Dal Peraro, M, Klein, M.L, Wand, A.J, DeGrado, W.F. | | Deposit date: | 2006-08-08 | | Release date: | 2007-07-17 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of a designed metalloprotein and complementary molecular dynamics refinement.
Structure, 16, 2008
|
|
5YGJ
 
 | | Crystal structure of a synthase from Streptomyces sp. CL190 | | Descriptor: | Cyclolavandulyl diphosphate synthase | | Authors: | Gao, J, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2017-09-23 | | Release date: | 2018-07-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.648 Å) | | Cite: | Catalytic Role of Conserved Asparagine, Glutamine, Serine, and Tyrosine Residues in Isoprenoid Biosynthesis Enzymes.
Acs Catalysis, 8, 2018
|
|
