8IBJ
 
 | | Inactive mutant of CtPL-H210S/F214I/N181A/F235L | | Descriptor: | PET hydrolase | | Authors: | Li, X, Shi, B.L, Zeng, Z.Y, Huang, J.-W, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2023-02-10 | | Release date: | 2023-04-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Functional tailoring of a PET hydrolytic enzyme expressed in Pichia pastoris.
Bioresour Bioprocess, 10, 2023
|
|
8IAN
 
 | | Crystal structure of CtPL-H210S/F214I mutant | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, PET hydrolase | | Authors: | Li, X, Shi, B.L, Zeng, Z.Y, Huang, J.-W, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2023-02-08 | | Release date: | 2023-04-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Functional tailoring of a PET hydrolytic enzyme expressed in Pichia pastoris.
Bioresour Bioprocess, 10, 2023
|
|
3CTZ
 
 | | Structure of human cytosolic X-prolyl aminopeptidase | | Descriptor: | CALCIUM ION, HEXAETHYLENE GLYCOL, MANGANESE (II) ION, ... | | Authors: | Li, X, Lou, Z, Rao, Z. | | Deposit date: | 2008-04-15 | | Release date: | 2008-05-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of human cytosolic X-prolyl aminopeptidase: a double Mn(II)-dependent dimeric enzyme with a novel three-domain subunit
J.Biol.Chem., 283, 2008
|
|
1KDU
 
 | | SEQUENTIAL 1H NMR ASSIGNMENTS AND SECONDARY STRUCTURE OF THE KRINGLE DOMAIN FROM UROKINASE | | Descriptor: | PLASMINOGEN ACTIVATOR | | Authors: | Li, X, Bokman, A.M, Llinas, M, Smith, R.A.G, Dobson, C.M. | | Deposit date: | 1993-07-15 | | Release date: | 1993-10-31 | | Last modified: | 2024-06-05 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the kringle domain from urokinase-type plasminogen activator.
J.Mol.Biol., 235, 1994
|
|
3PDV
 
 | |
1ZGK
 
 | | 1.35 angstrom structure of the Kelch domain of Keap1 | | Descriptor: | Kelch-like ECH-associated protein 1 | | Authors: | Li, X, Bottoms, C.A, Hannink, M, Beamer, L.J. | | Deposit date: | 2005-04-21 | | Release date: | 2005-10-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Conserved solvent and side-chain interactions in the 1.35 Angstrom structure of the Kelch domain of Keap1.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
5KWY
 
 | | Structure of human NPC1 middle lumenal domain bound to NPC2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLEST-5-EN-3-YL HYDROGEN SULFATE, Epididymal secretory protein E1, ... | | Authors: | Li, X. | | Deposit date: | 2016-07-19 | | Release date: | 2016-08-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.405 Å) | | Cite: | Clues to the mechanism of cholesterol transfer from the structure of NPC1 middle lumenal domain bound to NPC2.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
6VUM
 
 | |
3PYC
 
 | | Crystal structure of human SMURF1 C2 domain | | Descriptor: | CHLORIDE ION, E3 ubiquitin-protein ligase SMURF1, SULFATE ION | | Authors: | Li, X, Li, P. | | Deposit date: | 2010-12-13 | | Release date: | 2011-04-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal structure of human SMURF1 C2 domain
To be Published
|
|
1CS3
 
 | |
5EE9
 
 | | Complex structure of OSYCHF1 with GMP-PNP | | Descriptor: | GLYCEROL, MAGNESIUM ION, Obg-like ATPase 1, ... | | Authors: | Li, X, Chen, Z. | | Deposit date: | 2015-10-22 | | Release date: | 2016-02-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | ATP binding by the P-loop NTPase OsYchF1 (an unconventional G protein) contributes to biotic but not abiotic stress responses
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5EE1
 
 | | Crystal structure of OsYchF1 at pH 7.85 | | Descriptor: | Obg-like ATPase 1 | | Authors: | Li, X, Chen, Z. | | Deposit date: | 2015-10-22 | | Release date: | 2016-02-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | ATP binding by the P-loop NTPase OsYchF1 (an unconventional G protein) contributes to biotic but not abiotic stress responses
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5EE3
 
 | | COMPLEX STRUCTURE OF OSYCHF1 WITH AMP-PNP | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Li, X, Chen, Z. | | Deposit date: | 2015-10-22 | | Release date: | 2016-02-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | ATP binding by the P-loop NTPase OsYchF1 (an unconventional G protein) contributes to biotic but not abiotic stress responses
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
3T7J
 
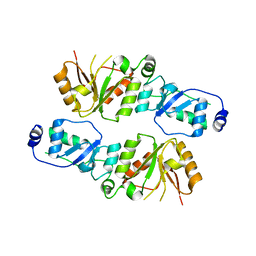 | | Crystal structure of Rtt107p (residues 820-1070) | | Descriptor: | Regulator of Ty1 transposition protein 107 | | Authors: | Li, X, Li, F, Wu, J, Shi, Y. | | Deposit date: | 2011-07-30 | | Release date: | 2012-02-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.042 Å) | | Cite: | Structure of C-terminal Tandem BRCT Repeats of Rtt107 Protein Reveals Critical Role in Interaction with Phosphorylated Histone H2A during DNA Damage Repair
J.Biol.Chem., 287, 2012
|
|
3T7I
 
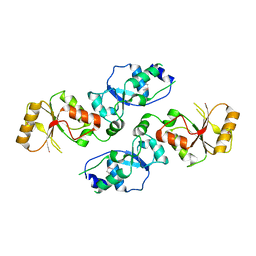 | | Crystal structure of Se-Met Rtt107p (residues 820-1070) | | Descriptor: | Regulator of Ty1 transposition protein 107 | | Authors: | Li, X, Li, F, Wu, J, Shi, Y. | | Deposit date: | 2011-07-30 | | Release date: | 2012-02-15 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of C-terminal Tandem BRCT Repeats of Rtt107 Protein Reveals Critical Role in Interaction with Phosphorylated Histone H2A during DNA Damage Repair
J.Biol.Chem., 287, 2012
|
|
3T7K
 
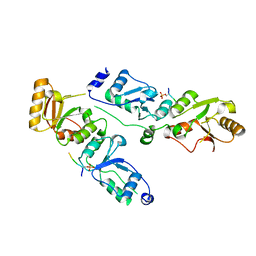 | | Complex structure of Rtt107p and phosphorylated histone H2A | | Descriptor: | Histone H2A.1, Regulator of Ty1 transposition protein 107 | | Authors: | Li, X, Li, F, Wu, J, Shi, Y. | | Deposit date: | 2011-07-30 | | Release date: | 2012-02-15 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.028 Å) | | Cite: | Structure of C-terminal Tandem BRCT Repeats of Rtt107 Protein Reveals Critical Role in Interaction with Phosphorylated Histone H2A during DNA Damage Repair
J.Biol.Chem., 287, 2012
|
|
2M9H
 
 | |
2FQD
 
 | | Crystal Structures of E. coli Laccase CueO under different copper binding situations | | Descriptor: | Blue copper oxidase cueO, CITRIC ACID, COPPER (II) ION, ... | | Authors: | Li, X, Wei, Z, Zhang, M, Teng, M, Gong, W. | | Deposit date: | 2006-01-18 | | Release date: | 2007-01-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of E. coli laccase CueO at different copper concentrations.
Biochem.Biophys.Res.Commun., 354, 2007
|
|
2FQF
 
 | | Crystal Structures of E. coli Laccase CueO under different copper binding situations | | Descriptor: | Blue copper oxidase cueO, CITRIC ACID, COPPER (II) ION, ... | | Authors: | Li, X, Wei, Z, Zhang, M, Teng, M, Gong, W. | | Deposit date: | 2006-01-18 | | Release date: | 2007-01-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of E. coli laccase CueO at different copper concentrations.
Biochem.Biophys.Res.Commun., 354, 2007
|
|
2FQE
 
 | | Crystal Structures of E. coli Laccase CueO under different copper binding situations | | Descriptor: | Blue copper oxidase cueO, CITRIC ACID, COPPER (II) ION, ... | | Authors: | Li, X, Wei, Z, Zhang, M, Teng, M, Gong, W. | | Deposit date: | 2006-01-18 | | Release date: | 2007-01-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal structures of E. coli laccase CueO at different copper concentrations.
Biochem.Biophys.Res.Commun., 354, 2007
|
|
2FQG
 
 | | Crystal Structures of E. coli Laccase CueO under different copper binding situations | | Descriptor: | Blue copper oxidase cueO, CITRIC ACID, COPPER (II) ION, ... | | Authors: | Li, X, Wei, Z, Zhang, M, Teng, M, Gong, W. | | Deposit date: | 2006-01-18 | | Release date: | 2007-01-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of E. coli laccase CueO at different copper concentrations.
Biochem.Biophys.Res.Commun., 354, 2007
|
|
1ZVF
 
 | | The crystal structure of 3-hydroxyanthranilate 3,4-dioxygenase from Saccharomyces cerevisiae | | Descriptor: | 3-hydroxyanthranilate 3,4-dioxygenase, NICKEL (II) ION | | Authors: | Li, X, Guo, M, Teng, M, Niu, L. | | Deposit date: | 2005-06-02 | | Release date: | 2006-06-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | The crystal structure of 3-hydroxyanthranilate 3,4-dioxygenase from Saccharomyces cerevisiae
To be published
|
|
4QRD
 
 | | Structure of Methionyl-tRNA Synthetase in complex with N-(1H-benzimidazol-2-ylmethyl)-N'-(2,4-dichlorophenyl)-6-(morpholin-4-yl)-1,3,5-triazine-2,4-diamine | | Descriptor: | MAGNESIUM ION, Methionyl-tRNA synthetase, N-(1H-benzimidazol-2-ylmethyl)-N'-(2,4-dichlorophenyl)-6-(morpholin-4-yl)-1,3,5-triazine-2,4-diamine | | Authors: | Li, X, Hilgers, M.T, Stidham, M, Brown-Driver, V, Shaw, K.J, Finn, J. | | Deposit date: | 2014-06-30 | | Release date: | 2015-07-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Discovery and SAR of a Novel Series of Pyrimidine Antibacterials Targeting Methionyl-tRNA Synthetase
to be published
|
|
4QUV
 
 | | Structure of an integral membrane delta(14)-sterol reductase | | Descriptor: | Delta(14)-sterol reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Li, X, Blobel, G. | | Deposit date: | 2014-07-12 | | Release date: | 2014-10-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.743 Å) | | Cite: | Structure of an integral membrane sterol reductase from Methylomicrobium alcaliphilum.
Nature, 517, 2015
|
|
3B9Y
 
 | | Crystal structure of the Nitrosomonas europaea Rh protein | | Descriptor: | Ammonium transporter family Rh-like protein, UNKNOWN LIGAND, octyl beta-D-glucopyranoside | | Authors: | Li, X, Jayachandran, S, Nguyen, H.-H.T, Chan, M.K. | | Deposit date: | 2007-11-07 | | Release date: | 2007-12-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of the Nitrosomonas europaea Rh protein.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
