4V2A
 
 | | human Unc5A ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, NETRIN RECEPTOR UNC5A | | Authors: | Seiradake, E, del Toro, D, Nagel, D, Cop, F, Haertl, R, Ruff, T, Seyit-Bremer, G, Harlos, K, Border, E.C, Acker-Palmer, A, Jones, E.Y, Klein, R. | | Deposit date: | 2014-10-08 | | Release date: | 2014-11-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Flrt Structure: Balancing Repulsion and Cell Adhesion in Cortical and Vascular Development
Neuron, 84, 2014
|
|
5NUZ
 
 | | Junin virus GP1 glycoprotein in complex with an antibody Fab fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ISOPROPYL ALCOHOL, ... | | Authors: | Zeltina, A, Krumm, S.A, Sahin, M, Struwe, W.B, Harlos, K, Nunberg, J.H, Crispin, M, Pinschewer, D.D, Doores, K.J, Bowden, T.A. | | Deposit date: | 2017-05-03 | | Release date: | 2017-06-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Convergent immunological solutions to Argentine hemorrhagic fever virus neutralization.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5MQW
 
 | | High-speed fixed-target serial virus crystallography | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Roedig, P, Ginn, H.M, Pakendorf, T, Sutton, G, Harlos, K, Walter, T.S, Meyer, J, Fischer, P, Duman, R, Vartiainen, I, Reime, B, Warmer, M, Brewster, A.S, Young, I.D, Michels-Clark, T, Sauter, N.K, Sikorsky, M, Nelson, S, Damiani, D.S, Alonso-Mori, R, Ren, J, Fry, E.E, David, C, Stuart, D.I, Wagner, A, Meents, A. | | Deposit date: | 2016-12-21 | | Release date: | 2017-06-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | High-speed fixed-target serial virus crystallography.
Nat. Methods, 14, 2017
|
|
5MGT
 
 | | Complex of human NKR-P1 and LLT1 in deglycosylated forms | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, C-type lectin domain family 2 member D, CHLORIDE ION, ... | | Authors: | Blaha, J, Skalova, T, Stransky, J, Koval, T, Hasek, J, Yuguang, Z, Harlos, K, Vanek, O, Dohnalek, J. | | Deposit date: | 2016-11-22 | | Release date: | 2018-06-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the human NK cell NKR-P1:LLT1 receptor:ligand complex reveals clustering in the immune synapse.
Nat Commun, 13, 2022
|
|
5MGS
 
 | | Human receptor NKR-P1 in deglycosylated form, extracellular domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Killer cell lectin-like receptor subfamily B member 1 | | Authors: | Skalova, T, Blaha, J, Stransky, J, Koval, T, Hasek, J, Yuguang, Z, Harlos, K, Vanek, O, Dohnalek, J. | | Deposit date: | 2016-11-22 | | Release date: | 2018-05-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the human NK cell NKR-P1:LLT1 receptor:ligand complex reveals clustering in the immune synapse.
Nat Commun, 13, 2022
|
|
6IPA
 
 | |
6QP7
 
 | | Drosophila Semaphorin 2a | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Robinson, R.A, Rozbesky, D, Harlos, K, Siebold, C, Jones, E.Y. | | Deposit date: | 2019-02-13 | | Release date: | 2019-08-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Diversity of oligomerization in Drosophila semaphorins suggests a mechanism of functional fine-tuning.
Nat Commun, 10, 2019
|
|
6QP8
 
 | | Drosophila Semaphorin 2b | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CITRATE ANION, ... | | Authors: | Robinson, R.A, Rozbesky, D, Harlos, K, Siebold, C, Jones, E.Y. | | Deposit date: | 2019-02-13 | | Release date: | 2019-08-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Diversity of oligomerization in Drosophila semaphorins suggests a mechanism of functional fine-tuning.
Nat Commun, 10, 2019
|
|
2CDG
 
 | | Structure and binding kinetics of three different human CD1d-alpha- Galactosylceramide-specific T cell receptors (TCR 5B) | | Descriptor: | TCR 5E | | Authors: | Gadola, S.D, Koch, M, Marles-Wright, J, Lissin, N.M, Sheperd, D, Matulis, G, Harlos, K, Villiger, P.M, Stuart, D.I, Jakobsen, B.K, Cerundolo, V, Jones, E.Y. | | Deposit date: | 2006-01-23 | | Release date: | 2006-03-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structrue and Binding Kinetics of Three Different Human Cd1D-Alpha-Galactosylceramide-Specific T Cell Receptors
J.Exp.Med., 203, 2006
|
|
2CDE
 
 | | Structure and binding kinetics of three different human CD1d-alpha- Galactosylceramide specific T cell receptors - iNKT-TCR | | Descriptor: | INKT-TCR | | Authors: | Gadola, S.D, Koch, M, Marles-Wright, J, Lissin, N.M, Sheperd, D, Matulis, G, Harlos, K, Villiger, P.M, Stuart, D.I, Jakobsen, B.K, Cerundolo, V, Jones, E.Y. | | Deposit date: | 2006-01-23 | | Release date: | 2006-03-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structrue and Binding Kinetics of Three Different Human Cd1D-Alpha-Galactosylceramide-Specific T Cell Receptors
J.Exp.Med., 203, 2006
|
|
6QP9
 
 | | Drosophila Semaphorin 1a, extracellular domains 1-2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Semaphorin-1A | | Authors: | Rozbesky, D, Robinson, R.A, Harlos, K, Siebold, C, Jones, E.Y. | | Deposit date: | 2019-02-13 | | Release date: | 2019-08-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Diversity of oligomerization in Drosophila semaphorins suggests a mechanism of functional fine-tuning.
Nat Commun, 10, 2019
|
|
2CDF
 
 | | Structure and binding kinetics of three different human CD1d-alpha- Galactosylceramide-specific T cell receptors (TCR 5E) | | Descriptor: | TCR 5E | | Authors: | Gadola, S.D, Koch, M, Marles-Wright, J, Lissin, N.M, Sheperd, D, Matulis, G, Harlos, K, Villiger, P.M, Stuart, D.I, Jakobsen, B.K, Cerundolo, V, Jones, E.Y. | | Deposit date: | 2006-01-23 | | Release date: | 2006-03-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structrue and Binding Kinetics of Three Different Human Cd1D-Alpha-Galactosylceramide-Specific T Cell Receptors
J.Exp.Med., 203, 2006
|
|
4V2C
 
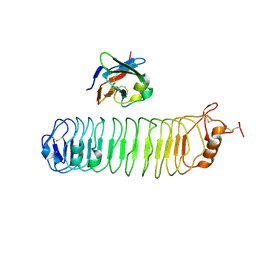 | | mouse FLRT2 LRR domain in complex with rat Unc5D Ig1 domain | | Descriptor: | FIBRONECTIN LEUCINE RICH TRANSMEMBRANE PROTEIN 2, PROTEIN UNC5D | | Authors: | Seiradake, E, del Toro, D, Nagel, D, Cop, F, Haertl, R, Ruff, T, Seyit-Bremer, G, Harlos, K, Border, E.C, Acker-Palmer, A, Jones, E.Y, Klein, R. | | Deposit date: | 2014-10-08 | | Release date: | 2014-11-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Flrt Structure: Balancing Repulsion and Cell Adhesion in Cortical and Vascular Development
Neuron, 84, 2014
|
|
4V2E
 
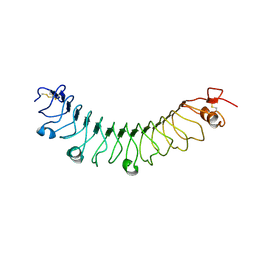 | | FLRT3 LRR domain | | Descriptor: | FIBRONECTIN LEUCINE RICH TRANSMEMBRANE PROTEIN 3 | | Authors: | Seiradake, E, del Toro, D, Nagel, D, Cop, F, Haertl, R, Ruff, T, Seyit-Bremer, G, Harlos, K, Border, E.C, Acker-Palmer, A, Jones, E.Y, Klein, R. | | Deposit date: | 2014-10-08 | | Release date: | 2014-11-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Flrt Structure: Balancing Repulsion and Cell Adhesion in Cortical and Vascular Development.
Neuron, 84, 2014
|
|
5MGR
 
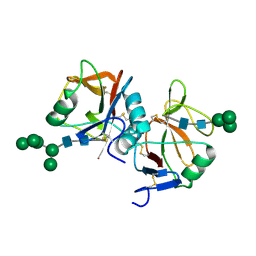 | | Human receptor NKR-P1 in glycosylated form, extracellular domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Killer cell lectin-like receptor subfamily B member 1, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Skalova, T, Blaha, J, Stransky, J, Koval, T, Hasek, J, Yuguang, Z, Harlos, K, Vanek, O, Dohnalek, J. | | Deposit date: | 2016-11-22 | | Release date: | 2018-06-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the human NK cell NKR-P1:LLT1 receptor:ligand complex reveals clustering in the immune synapse.
Nat Commun, 13, 2022
|
|
4WK9
 
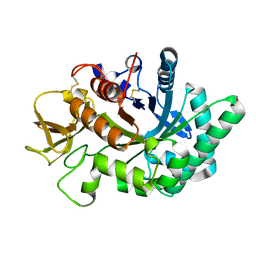 | | Crystal structure of human chitotriosidase-1 catalytic domain in complex with chitobiose (0.3mM) at 1.10 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitotriosidase-1 | | Authors: | Fadel, F, Zhao, Y, Cachau, R, Cousido-Siah, A, Ruiz, F.X, Harlos, K, Howard, E, Mitschler, A, Podjarny, A. | | Deposit date: | 2014-10-02 | | Release date: | 2015-07-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.102 Å) | | Cite: | New insights into the enzymatic mechanism of human chitotriosidase (CHIT1) catalytic domain by atomic resolution X-ray diffraction and hybrid QM/MM.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4WJX
 
 | | Crystal structure of human chitotriosidase-1 catalytic domain at 1.0 A resolution | | Descriptor: | Chitotriosidase-1, L(+)-TARTARIC ACID | | Authors: | Fadel, F, Zhao, Y, Cachau, R, Cousido-Siah, A, Ruiz, F.X, Harlos, K, Howard, E, Mitschler, A, Podjarny, A. | | Deposit date: | 2014-10-01 | | Release date: | 2015-07-08 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | New insights into the enzymatic mechanism of human chitotriosidase (CHIT1) catalytic domain by atomic resolution X-ray diffraction and hybrid QM/MM
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4WKH
 
 | | Crystal structure of human chitotriosidase-1 catalytic domain in complex with chitobiose (1mM) at 1.05 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitotriosidase-1 | | Authors: | Fadel, F, Zhao, Y, Cachau, R, Cousido-Siah, A, Ruiz, F.X, Harlos, K, Howard, E, Mitschler, A, Podjarny, A. | | Deposit date: | 2014-10-02 | | Release date: | 2015-07-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | New insights into the enzymatic mechanism of human chitotriosidase (CHIT1) catalytic domain by atomic resolution X-ray diffraction and hybrid QM/MM.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4WKF
 
 | | Crystal structure of human chitotriosidase-1 catalytic domain in complex with chitobiose (2.5mM) at 1.10 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitotriosidase-1 | | Authors: | Fadel, F, Zhao, Y, Cachau, R, Cousido-Siah, A, Ruiz, F.X, Harlos, K, Howard, E, Mitschler, A, Podjarny, A. | | Deposit date: | 2014-10-02 | | Release date: | 2015-07-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.101 Å) | | Cite: | New insights into the enzymatic mechanism of human chitotriosidase (CHIT1) catalytic domain by atomic resolution X-ray diffraction and hybrid QM/MM.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4WKA
 
 | | Crystal structure of human chitotriosidase-1 catalytic domain at 0.95 A resolution | | Descriptor: | Chitotriosidase-1, L(+)-TARTARIC ACID | | Authors: | Fadel, F, Zhao, Y, Cachau, R, Cousido-Siah, A, Ruiz, F.X, Harlos, K, Howard, E, Mitschler, A, Podjarny, A. | | Deposit date: | 2014-10-02 | | Release date: | 2015-07-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | New insights into the enzymatic mechanism of human chitotriosidase (CHIT1) catalytic domain by atomic resolution X-ray diffraction and hybrid QM/MM.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
2BRY
 
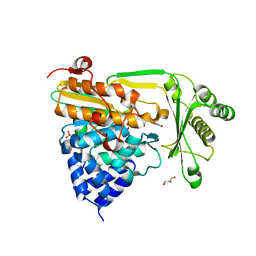 | | Crystal structure of the native monooxygenase domain of MICAL at 1.45 A resolution | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Siebold, C, Berrow, N, Walter, T.S, Harlos, K, Owens, R.J, Terman, J.R, Stuart, D.I, Kolodkin, A.L, Pasterkamp, R.J, Jones, E.Y. | | Deposit date: | 2005-05-13 | | Release date: | 2005-10-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | High-Resolution Structure of the Catalytic Region of Mical (Molecule Interacting with Casl), a Multidomain Flavoenzyme-Signaling Molecule.
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
1VCA
 
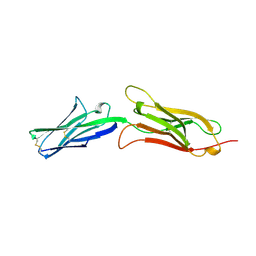 | | CRYSTAL STRUCTURE OF AN INTEGRIN-BINDING FRAGMENT OF VASCULAR CELL ADHESION MOLECULE-1 AT 1.8 ANGSTROMS RESOLUTION | | Descriptor: | HUMAN VASCULAR CELL ADHESION MOLECULE-1 | | Authors: | Jones, E.Y, Harlos, K, Bottomley, M.J, Robinson, R.C, Driscoll, P.C, Edwards, R.M, Clements, J.M, Dudgeon, T.J, Stuart, D.I. | | Deposit date: | 1995-03-21 | | Release date: | 1995-09-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of an integrin-binding fragment of vascular cell adhesion molecule-1 at 1.8 A resolution.
Nature, 373, 1995
|
|
6JRE
 
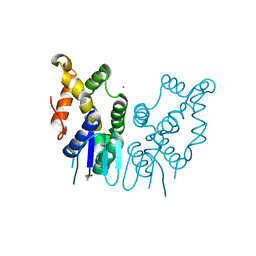 | | Structure of N-terminal domain of Plasmodium vivax p43 (PfNTD) solved by Co-SAD phasing | | Descriptor: | Aminoacyl-tRNA synthetase-interacting multifunctional protein p43, COBALT (II) ION | | Authors: | Manickam, Y, Harlos, K, Sharma, M, Gupta, S, Sharma, A. | | Deposit date: | 2019-04-03 | | Release date: | 2020-03-11 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Crystal structures of the two domains that constitute the Plasmodium vivax p43 protein.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
1OLZ
 
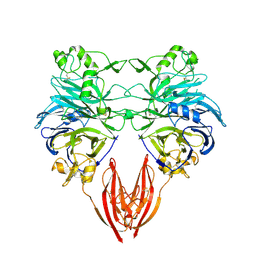 | | The ligand-binding face of the semaphorins revealed by the high resolution crystal structure of SEMA4D | | Descriptor: | SEMAPHORIN 4D | | Authors: | Love, C.A, Harlos, K, Mavaddat, N, Davis, S.J, Stuart, D.I, Jones, E.Y, Esnouf, R.M. | | Deposit date: | 2003-08-19 | | Release date: | 2003-09-11 | | Last modified: | 2018-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Ligand-Binding Face of the Semaphorins Revealed by the High-Resolution Crystal Structure of Sema4D
Nat.Struct.Biol., 10, 2003
|
|
1QFV
 
 | | HISTAMINE BINDING PROTEIN FROM FEMALE BROWN EAR RHIPICEPHALUS APPENDICULATUS | | Descriptor: | HISTAMINE, PROTEIN (FEMALE-SPECIFIC HISTAMINE BINDING PROTEIN 2) | | Authors: | Paesen, G.C, Adams, P.L, Harlos, K, Nuttal, P.A, Stuart, D.I. | | Deposit date: | 1999-04-14 | | Release date: | 2000-04-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Tick histamine-binding proteins: isolation, cloning, and three-dimensional structure.
Mol.Cell, 3, 1999
|
|
