5EJ6
 
 | | EcMenD-ThDP-Mn2+ complex soaked with 2-ketoglutarate for 2min then soaked with isochorismate for 2 min | | Descriptor: | (4S)-4-{3-[(4-amino-2-methylpyrimidin-5-yl)methyl]-5-(2-{[(S)-hydroxy(phosphonooxy)phosphoryl]oxy}ethyl)-4-methyl-1,3lambda~5~-thiazol-2-yl}-4-hydroxybutanoic acid, 2-succinyl-5-enolpyruvyl-6-hydroxy-3-cyclohexene-1-carboxylate synthase, MANGANESE (II) ION | | Authors: | Song, H.G, Dong, C, Chen, Y.Z, Sun, Y.R, Guo, Z.H. | | Deposit date: | 2015-11-01 | | Release date: | 2016-06-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.243 Å) | | Cite: | A Thiamine-Dependent Enzyme Utilizes an Active Tetrahedral Intermediate in Vitamin K Biosynthesis
J.Am.Chem.Soc., 138, 2016
|
|
5EJ4
 
 | | EcMenD-ThDP-Mn2+ complex soaked with 2-ketoglutarate for 15 min | | Descriptor: | (4S)-4-{3-[(4-amino-2-methylpyrimidin-5-yl)methyl]-5-(2-{[(S)-hydroxy(phosphonooxy)phosphoryl]oxy}ethyl)-4-methyl-1,3lambda~5~-thiazol-2-yl}-4-hydroxybutanoic acid, 2-succinyl-5-enolpyruvyl-6-hydroxy-3-cyclohexene-1-carboxylate synthase, FORMIC ACID, ... | | Authors: | Song, H.G, Dong, C, Chen, Y.Z, Sun, Y.R, Guo, Z.H. | | Deposit date: | 2015-11-01 | | Release date: | 2016-06-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.773 Å) | | Cite: | A Thiamine-Dependent Enzyme Utilizes an Active Tetrahedral Intermediate in Vitamin K Biosynthesis
J.Am.Chem.Soc., 138, 2016
|
|
5EJ8
 
 | | EcMenD-ThDP-Mn2+ complex structure soaked with 2-ketoglutarate for 2 min | | Descriptor: | (4S)-4-{3-[(4-amino-2-methylpyrimidin-5-yl)methyl]-5-(2-{[(S)-hydroxy(phosphonooxy)phosphoryl]oxy}ethyl)-4-methyl-1,3lambda~5~-thiazol-2-yl}-4-hydroxybutanoic acid, 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Song, H.G, Dong, C, Chen, Y.Z, Sun, Y.R, Guo, Z.H. | | Deposit date: | 2015-11-01 | | Release date: | 2016-06-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | A Thiamine-Dependent Enzyme Utilizes an Active Tetrahedral Intermediate in Vitamin K Biosynthesis
J.Am.Chem.Soc., 138, 2016
|
|
5A9V
 
 | | Structure of apo BipA | | Descriptor: | GTP-BINDING PROTEIN | | Authors: | Kumar, V, Chen, Y, Ero, R, Li, Z, Gao, Y. | | Deposit date: | 2015-07-23 | | Release date: | 2015-09-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | Structure of Bipa in GTP Form Bound to the Ratcheted Ribosome.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
2ASQ
 
 | | Solution Structure of SUMO-1 in Complex with a SUMO-binding Motif (SBM) | | Descriptor: | Protein inhibitor of activated STAT2, Small ubiquitin-related modifier 1 | | Authors: | Song, J, Zhang, Z, Hu, W, Chen, Y. | | Deposit date: | 2005-08-23 | | Release date: | 2005-10-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Small Ubiquitin-like Modifier (SUMO) Recognition of a SUMO Binding Motif: A reversal of the bound orientation
J.Biol.Chem., 280, 2005
|
|
1YPX
 
 | | Crystal Structure of the Putative Vitamin-B12 Independent Methionine Synthase from Listeria monocytogenes, Northeast Structural Genomics Target LmR13 | | Descriptor: | PHOSPHATE ION, putative vitamin-B12 independent methionine synthase family protein | | Authors: | Forouhar, F, Chen, Y, Benach, J, Xiao, R, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-01-31 | | Release date: | 2005-02-08 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of Putative Vitamin-B12 Independent Methionine Synthase from Listeria monocytogenes, Northeast Structural Genomics Target LmR13
To be Published
|
|
4NES
 
 | | Crystal structure of Methanocaldococcus jannaschii UDP-GlcNAc 2-epimerase in complex with UDP-GlcNAc and UDP | | Descriptor: | UDP-N-acetylglucosamine 2-epimerase, URIDINE-5'-DIPHOSPHATE, URIDINE-DIPHOSPHATE-N-ACETYLGLUCOSAMINE | | Authors: | Chen, S.C, Yang, C.S, Huang, C.H, Chen, Y. | | Deposit date: | 2013-10-30 | | Release date: | 2014-04-23 | | Last modified: | 2015-06-17 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Crystal structures of the archaeal UDP-GlcNAc 2-epimerase from Methanocaldococcus jannaschii reveal a conformational change induced by UDP-GlcNAc.
Proteins, 82, 2014
|
|
5A9W
 
 | | Structure of GDPCP BipA | | Descriptor: | GTP-BINDING PROTEIN, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER | | Authors: | Kumar, V, Chen, Y, Ero, R, Li, Z, Gao, Y. | | Deposit date: | 2015-07-23 | | Release date: | 2015-08-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structure of Bipa in GTP Form Bound to the Ratcheted Ribosome.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
2IE4
 
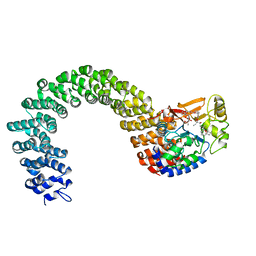 | | Structure of the Protein Phosphatase 2A Core Enzyme Bound to okadaic acid | | Descriptor: | MANGANESE (II) ION, OKADAIC ACID, Protein Phosphatase 2, ... | | Authors: | Xing, Y, Xu, Y, Chen, Y, Jeffrey, P.D, Chao, Y, Shi, Y. | | Deposit date: | 2006-09-17 | | Release date: | 2006-11-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of Protein Phosphatase 2A Core Enzyme Bound to Tumor-Inducing Toxins
Cell(Cambridge,Mass.), 127, 2006
|
|
5EJ9
 
 | | EcMenD-ThDP-Mn2+ complex soaked with 2-ketoglutarate for 2 min and isochorismate for 13 min | | Descriptor: | 2-succinyl-5-enolpyruvyl-6-hydroxy-3-cyclohexene-1-carboxylate synthase, FORMIC ACID, GLYCEROL, ... | | Authors: | Song, H.G, Dong, C, Chen, Y.Z, Sun, Y.R, Guo, Z.H. | | Deposit date: | 2015-11-01 | | Release date: | 2016-06-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | A Thiamine-Dependent Enzyme Utilizes an Active Tetrahedral Intermediate in Vitamin K Biosynthesis
J.Am.Chem.Soc., 138, 2016
|
|
3V36
 
 | | Aldose reductase complexed with glceraldehyde | | Descriptor: | Aldose reductase, D-Glyceraldehyde, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Zheng, X, Zhang, L, Chen, Y, Luo, H, Hu, X. | | Deposit date: | 2011-12-13 | | Release date: | 2012-08-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Partial inhibition of aldose reductase by nitazoxanide and its molecular basis.
Chemmedchem, 7, 2012
|
|
2B6E
 
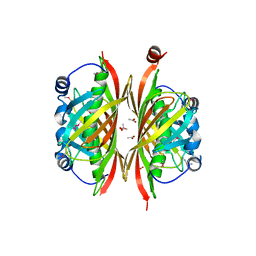 | | X-Ray Crystal Structure of Protein HI1161 from Haemophilus influenzae. Northeast Structural Genomics Consortium Target IR63. | | Descriptor: | ACETIC ACID, Hypothetical UPF0152 protein HI1161 | | Authors: | Kuzin, A.P, Benach, J, Chen, Y, Acton, T, Xiao, R, Conover, K, Ma, L.-C, Kellie, R, Cunningham, K.E, Montelione, G, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-10-01 | | Release date: | 2005-10-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-Ray structure of the different crystal form of the hypothetical UPF0152 protein HI1161: NESG target IR63
TO BE PUBLISHED
|
|
2AJA
 
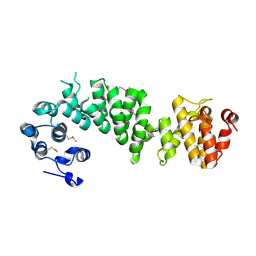 | | X-Ray structure of an ankyrin repeat family protein Q5ZSV0 from Legionella pneumophila. Northeast Structural Genomics Consortium target LgR21. | | Descriptor: | ankyrin repeat family protein | | Authors: | Kuzin, A.P, Chen, Y, Acton, T, Xiao, R, Conover, K, Ma, C, Kellie, R, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-08-01 | | Release date: | 2005-08-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-Ray structure of an ankyrin repeat family protein Q5ZSV0 from Legionella pneumophila.
To be Published
|
|
4NEQ
 
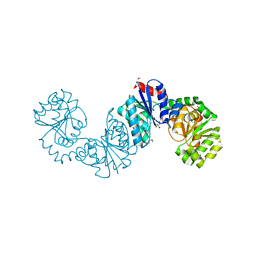 | | The structure of UDP-GlcNAc 2-epimerase from Methanocaldococcus jannaschii | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, UDP-N-acetylglucosamine 2-epimerase | | Authors: | Chen, S.C, Yang, C.S, Huang, C.H, Chen, Y. | | Deposit date: | 2013-10-29 | | Release date: | 2014-04-23 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal structures of the archaeal UDP-GlcNAc 2-epimerase from Methanocaldococcus jannaschii reveal a conformational change induced by UDP-GlcNAc.
Proteins, 82, 2014
|
|
5EJ7
 
 | | EcMenD-ThDP-Mn2+ complex soaked with 2-ketoglutarate for 21 s | | Descriptor: | (4S)-4-{3-[(4-amino-2-methylpyrimidin-5-yl)methyl]-5-(2-{[(S)-hydroxy(phosphonooxy)phosphoryl]oxy}ethyl)-4-methyl-1,3lambda~5~-thiazol-2-yl}-4-hydroxybutanoic acid, 2-succinyl-5-enolpyruvyl-6-hydroxy-3-cyclohexene-1-carboxylate synthase, GLYCEROL, ... | | Authors: | Song, H.G, Dong, C, Chen, Y.Z, Sun, Y.R, Guo, Z.H. | | Deposit date: | 2015-11-01 | | Release date: | 2016-06-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | A Thiamine-Dependent Enzyme Utilizes an Active Tetrahedral Intermediate in Vitamin K Biosynthesis
J.Am.Chem.Soc., 138, 2016
|
|
5EJ5
 
 | | EcMenD-ThDP-Mn2+ complex soaked with 2-ketoglutarate for 1.5 h | | Descriptor: | (4S)-4-{3-[(4-amino-2-methylpyrimidin-5-yl)methyl]-5-(2-{[(S)-hydroxy(phosphonooxy)phosphoryl]oxy}ethyl)-4-methyl-1,3lambda~5~-thiazol-2-yl}-4-hydroxybutanoic acid, 2-succinyl-5-enolpyruvyl-6-hydroxy-3-cyclohexene-1-carboxylate synthase, GLYCEROL, ... | | Authors: | Song, H.G, Dong, C, Chen, Y.Z, Sun, Y.R, Guo, Z.H. | | Deposit date: | 2015-11-01 | | Release date: | 2016-06-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A Thiamine-Dependent Enzyme Utilizes an Active Tetrahedral Intermediate in Vitamin K Biosynthesis
J.Am.Chem.Soc., 138, 2016
|
|
5EJA
 
 | | EcMenD-ThDP-Mn2+ complex soaked with 2-ketoglutarate for 2 min and soaked with isochorismate for 7 min | | Descriptor: | (4S)-4-{3-[(4-amino-2-methylpyrimidin-5-yl)methyl]-5-(2-{[(S)-hydroxy(phosphonooxy)phosphoryl]oxy}ethyl)-4-methyl-1,3lambda~5~-thiazol-2-yl}-4-hydroxybutanoic acid, 2-succinyl-5-enolpyruvyl-6-hydroxy-3-cyclohexene-1-carboxylate synthase, GLYCEROL, ... | | Authors: | Song, H.G, Dong, C, Chen, Y.Z, Sun, Y.R, Guo, Z.H. | | Deposit date: | 2015-11-01 | | Release date: | 2016-06-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A Thiamine-Dependent Enzyme Utilizes an Active Tetrahedral Intermediate in Vitamin K Biosynthesis
J.Am.Chem.Soc., 138, 2016
|
|
5GOP
 
 | | Crystal structure of alkaline invertase InvA from Anabaena sp. PCC 7120 complexed with sucrose | | Descriptor: | Alkaline Invertase, beta-D-fructofuranose, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose | | Authors: | Xie, J, Cai, K, Hu, H.X, Jiang, Y.L, Yang, F, Hu, P.F, Chen, Y, Zhou, C.Z. | | Deposit date: | 2016-07-28 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural Analysis of the Catalytic Mechanism and Substrate Specificity of Anabaena Alkaline Invertase InvA Reveals a Novel Glucosidase
J. Biol. Chem., 291, 2016
|
|
2NPP
 
 | | Structure of the Protein Phosphatase 2A Holoenzyme | | Descriptor: | MANGANESE (II) ION, Protein Phosphatase 2, regulatory subunit A (PR 65), ... | | Authors: | Xu, Y, Chen, Y, Xing, Y, Chao, Y, Shi, Y. | | Deposit date: | 2006-10-28 | | Release date: | 2006-12-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of the protein phosphatase 2A holoenzyme
Cell(Cambridge,Mass.), 127, 2006
|
|
1ZBR
 
 | | Crystal Structure of the Putative Arginine Deiminase from Porphyromonas gingivalis, Northeast Structural Genomics Target PgR3 | | Descriptor: | conserved hypothetical protein | | Authors: | Forouhar, F, Chen, Y, Kuzin, A, Conover, K, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-04-08 | | Release date: | 2005-04-19 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of the Putative Arginine Deiminase from Porphyromonas gingivalis, Northeast Structural Genomics Target PgR3
To be Published
|
|
5GOQ
 
 | | Crystal structure of alkaline invertase InvA from Anabaena sp. PCC 7120 complexed with glucose | | Descriptor: | Alkaline Invertase, alpha-D-glucopyranose | | Authors: | Xie, J, Cai, K, Hu, H.X, Jiang, Y.L, Yang, F, Hu, P.F, Chen, Y, Zhou, C.Z. | | Deposit date: | 2016-07-28 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural Analysis of the Catalytic Mechanism and Substrate Specificity of Anabaena Alkaline Invertase InvA Reveals a Novel Glucosidase
J. Biol. Chem., 291, 2016
|
|
5GOO
 
 | | Crystal structure of alkaline invertase InvA from Anabaena sp. PCC 7120 complexed with fructose | | Descriptor: | Alkaline Invertase, GLYCEROL, beta-D-fructofuranose | | Authors: | Xie, J, Cai, K, Hu, H.X, Jiang, Y.L, Yang, F, Hu, P.F, Chen, Y, Zhou, C.Z. | | Deposit date: | 2016-07-28 | | Release date: | 2016-11-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structural Analysis of the Catalytic Mechanism and Substrate Specificity of Anabaena Alkaline Invertase InvA Reveals a Novel Glucosidase.
J. Biol. Chem., 291, 2016
|
|
2ATG
 
 | | NMR structure of Retrocyclin-2 in SDS | | Descriptor: | Retrocyclin-2 | | Authors: | Daly, N.L, Chen, Y.K, Rosengren, K.J, Marx, U.C, Phillips, M.L, Waring, A.J, Wang, W, Lehrer, R.I, Craik, D.J. | | Deposit date: | 2005-08-24 | | Release date: | 2005-09-06 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Retrocyclin-2: structural analysis of a potent anti-HIV theta-defensin
Biochemistry, 46, 2007
|
|
2NYL
 
 | | Crystal structure of Protein Phosphatase 2A (PP2A) holoenzyme with the catalytic subunit carboxyl terminus truncated | | Descriptor: | MANGANESE (II) ION, Protein phosphatase 2, regulatory subunit A (PR 65), ... | | Authors: | Xing, Y, Xu, Y, Chen, Y, Chao, Y, Lin, Z, Shi, Y. | | Deposit date: | 2006-11-20 | | Release date: | 2006-12-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structure of the Protein Phosphatase 2A Holoenzyme.
Cell(Cambridge,Mass.), 127, 2006
|
|
3UBI
 
 | | The Absence of Tertiary Interactions in a Self-Assembled DNA Crystal Structure | | Descriptor: | DNA (5'-D(*GP*AP*GP*CP*AP*GP*CP*CP*CP*GP*TP*AP*CP*TP*CP*G)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*TP*GP*AP*GP*GP*CP*TP*GP*C)-3'), DNA (5'-D(P*CP*CP*GP*AP*GP*TP*AP*CP*GP*AP*CP*GP*AP*CP*AP*AP*G)-3'), ... | | Authors: | Nguyen, N, Birktoft, J.J, Sha, R, Wang, T, Zheng, J, Constantinou, P.E, Ginell, S.L, Chen, Y, Mao, C, Seeman, N.C. | | Deposit date: | 2011-10-24 | | Release date: | 2012-05-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (6.8046 Å) | | Cite: | The absence of tertiary interactions in a self-assembled DNA crystal structure.
J.Mol.Recognit., 25, 2012
|
|
