4Z0L
 
 | | The murine cyclooxygenase-2 complexed with a nido-dicarbaborate-containing indomethacin derivative | | 分子名称: | (R)-7-{[5-methoxy-2-methyl-3-(methoxycarbonylmethyl)-1H-indolyl]carbonyl}-7,8-dicarba-nido-dodeca-hydroundecaborate, (S)-7-{[5-methoxy-2-methyl-3-(methoxycarbonylmethyl)-1H-indolyl]carbonyl}-7,8-dicarba-nido-dodeca-hydroundecaborate, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Xu, S, Neumann, W, Banerjee, S, Hey-Hawkins, E, Marnett, L.J. | | 登録日 | 2015-03-26 | | 公開日 | 2015-06-10 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.29 Å) | | 主引用文献 | nido-Dicarbaborate Induces Potent and Selective Inhibition of Cyclooxygenase-2.
Chemmedchem, 11, 2016
|
|
6X6O
 
 | | Crystal structure of T4 protein Spackle as determined by native SAD phasing | | 分子名称: | CHLORIDE ION, Protein spackle | | 著者 | Shi, K, Kurniawan, F, Banerjee, S, Moeller, N.H, Aihara, H. | | 登録日 | 2020-05-28 | | 公開日 | 2020-09-16 | | 実験手法 | X-RAY DIFFRACTION (1.52 Å) | | 主引用文献 | Crystal structure of bacteriophage T4 Spackle as determined by native SAD phasing.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
8B9X
 
 | | Chimeric protein of human UFM1 E3 ligase, UFL1, and DDRGK1 | | 分子名称: | CHLORIDE ION, DDRGK domain-containing protein 1,E3 UFM1-protein ligase 1 | | 著者 | Wiener, R, Isupov, M, banerjee, S. | | 登録日 | 2022-10-10 | | 公開日 | 2023-10-25 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (3.066 Å) | | 主引用文献 | Structural study of UFL1-UFC1 interaction uncovers the role of UFL1 N-terminal helix in ufmylation.
Embo Rep., 24, 2023
|
|
6E5T
 
 | | Crystal structure of human cellular retinol binding protein 1 in complex with abnormal-cannabidiorcin (Abn-CBDO) | | 分子名称: | (1'R,2'R)-5',6-dimethyl-2'-(prop-1-en-2-yl)-1',2',3',4'-tetrahydro[1,1'-biphenyl]-2,4-diol, Retinol-binding protein 1 | | 著者 | Silvaroli, J.A, Horwitz, S, Banerjee, S, Kiser, P.D, Golczak, M. | | 登録日 | 2018-07-23 | | 公開日 | 2019-02-13 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Abnormal Cannabidiol Modulates Vitamin A Metabolism by Acting as a Competitive Inhibitor of CRBP1.
Acs Chem.Biol., 14, 2019
|
|
6E6M
 
 | | Crystal structure of human cellular retinol-binding protein 1 in complex with cannabidiorcin (CBDO) | | 分子名称: | (1'R,2'R)-4,5'-dimethyl-2'-(prop-1-en-2-yl)-1',2',3',4'-tetrahydro[1,1'-biphenyl]-2,6-diol, Retinol-binding protein 1 | | 著者 | Silvaroli, J.A, Horwitz, S, Banerjee, S, Kiser, P.D, Golczak, M. | | 登録日 | 2018-07-25 | | 公開日 | 2019-02-13 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Abnormal Cannabidiol Modulates Vitamin A Metabolism by Acting as a Competitive Inhibitor of CRBP1.
Acs Chem.Biol., 14, 2019
|
|
4EBC
 
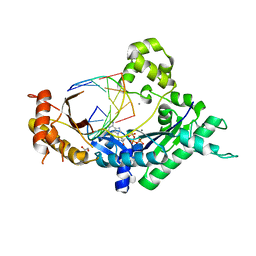 | | Conformationally Restrained North-methanocarba-2'-deoxyadenosine Corrects the Error-Prone Nature of Human DNA Polymerase Iota | | 分子名称: | 5'-D(P*AP*GP*GP*AP*CP*CP*(DOC))-3', 5'-D(P*CP*TP*GP*GP*GP*TP*CP*CP*T)-3', CALCIUM ION, ... | | 著者 | Eoff, R.L, Ketkar, A, Banerjee, S. | | 登録日 | 2012-03-23 | | 公開日 | 2012-06-13 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.901 Å) | | 主引用文献 | A Nucleotide-Analogue-Induced Gain of Function Corrects the Error-Prone Nature of Human DNA Polymerase iota.
J.Am.Chem.Soc., 134, 2012
|
|
4EBD
 
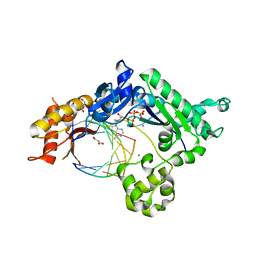 | | Conformationally Restrained North-methanocarba-2'-deoxyadenosine Corrects the Error-Prone Nature of Human DNA Polymerase Iota | | 分子名称: | 5'-D(P*AP*GP*GP*AP*CP*CP*(DOC))-3', 5'-D(P*CP*TP*GP*GP*GP*TP*CP*CP*T)-3', CALCIUM ION, ... | | 著者 | Eoff, R.L, Ketkar, A, Banerjee, S. | | 登録日 | 2012-03-23 | | 公開日 | 2012-06-13 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.571 Å) | | 主引用文献 | A Nucleotide-Analogue-Induced Gain of Function Corrects the Error-Prone Nature of Human DNA Polymerase iota.
J.Am.Chem.Soc., 134, 2012
|
|
4EBE
 
 | | Conformationally Restrained North-methanocarba-2'-deoxyadenosine Corrects the Error-Prone Nature of Human DNA Polymerase Iota | | 分子名称: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, 5'-D(P*AP*GP*GP*AP*CP*CP*(DOC))-3', 5'-D(P*CP*TP*GP*GP*GP*TP*CP*CP*T)-3', ... | | 著者 | Eoff, R.L, Ketkar, A, Banerjee, S. | | 登録日 | 2012-03-23 | | 公開日 | 2012-06-13 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | A Nucleotide-Analogue-Induced Gain of Function Corrects the Error-Prone Nature of Human DNA Polymerase iota.
J.Am.Chem.Soc., 134, 2012
|
|
6E5L
 
 | | Crystal structure of human cellular retinol binding protein 1 in complex with abnormal-cannabidiol (abn-CBD) | | 分子名称: | (1'R,2'R)-5'-methyl-6-pentyl-2'-(prop-1-en-2-yl)-1',2',3',4'-tetrahydro[1,1'-biphenyl]-2,4-diol, Retinol-binding protein 1 | | 著者 | Silvaroli, J.A, Banerjee, S, Kiser, P.D, Golczak, M. | | 登録日 | 2018-07-20 | | 公開日 | 2019-02-13 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.17 Å) | | 主引用文献 | Abnormal Cannabidiol Modulates Vitamin A Metabolism by Acting as a Competitive Inhibitor of CRBP1.
Acs Chem.Biol., 14, 2019
|
|
6E5W
 
 | | Crystal structure of human cellular retinol binding protein 3 in complex with abnormal-cannabidiol (abn-CBD) | | 分子名称: | (1'R,2'R)-5'-methyl-6-pentyl-2'-(prop-1-en-2-yl)-1',2',3',4'-tetrahydro[1,1'-biphenyl]-2,4-diol, GLYCEROL, Retinol-binding protein 5 | | 著者 | Silvaroli, J.A, Banerjee, S, Kiser, P.D, Golczak, M. | | 登録日 | 2018-07-23 | | 公開日 | 2019-02-13 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Abnormal Cannabidiol Modulates Vitamin A Metabolism by Acting as a Competitive Inhibitor of CRBP1.
Acs Chem.Biol., 14, 2019
|
|
6E6K
 
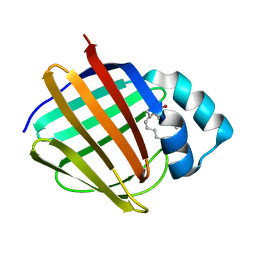 | | Crystal structure of human cellular retinol-binding protein 4 in complex with abnormal-cannabidiol (abn-CBD) | | 分子名称: | (1'R,2'R)-5'-methyl-6-pentyl-2'-(prop-1-en-2-yl)-1',2',3',4'-tetrahydro[1,1'-biphenyl]-2,4-diol, Retinoid-binding protein 7 | | 著者 | Silvaroli, J.A, Banerjee, S, Kiser, P.D, Golczak, M. | | 登録日 | 2018-07-25 | | 公開日 | 2019-02-13 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Abnormal Cannabidiol Modulates Vitamin A Metabolism by Acting as a Competitive Inhibitor of CRBP1.
Acs Chem.Biol., 14, 2019
|
|
6P7A
 
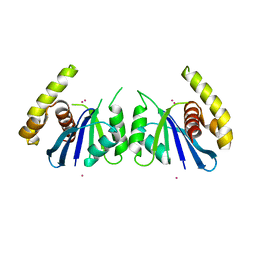 | | CRYSTAL STRUCTURE OF THE FOWLPOX VIRUS HOLLIDAY JUNCTION RESOLVASE | | 分子名称: | CADMIUM ION, Holliday junction resolvase | | 著者 | Li, N, Shi, K, Banerjee, S, Rao, T, Aihara, H. | | 登録日 | 2019-06-05 | | 公開日 | 2020-04-29 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (3.081 Å) | | 主引用文献 | Structural insights into the promiscuous DNA binding and broad substrate selectivity of fowlpox virus resolvase.
Sci Rep, 10, 2020
|
|
6P7B
 
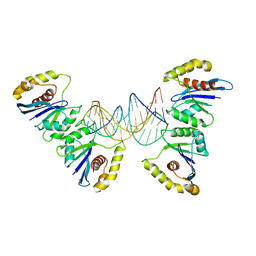 | | Crystal structure of Fowlpox virus resolvase and substrate Holliday junction DNA complex | | 分子名称: | DNA (29-MER), Holliday junction resolvase | | 著者 | Li, N, Shi, K, Rao, T, Banerjee, S, Aihara, H. | | 登録日 | 2019-06-05 | | 公開日 | 2020-04-29 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (3.317 Å) | | 主引用文献 | Structural insights into the promiscuous DNA binding and broad substrate selectivity of fowlpox virus resolvase.
Sci Rep, 10, 2020
|
|
3JD0
 
 | | Glutamate dehydrogenase in complex with GTP | | 分子名称: | GUANOSINE-5'-TRIPHOSPHATE, Glutamate dehydrogenase 1, mitochondrial | | 著者 | Borgnia, M.J, Banerjee, S, Merk, A, Matthies, D, Bartesaghi, A, Rao, P, Pierson, J, Earl, L.A, Falconieri, V, Subramaniam, S, Milne, J.L.S. | | 登録日 | 2016-03-28 | | 公開日 | 2016-04-27 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (3.47 Å) | | 主引用文献 | Using Cryo-EM to Map Small Ligands on Dynamic Metabolic Enzymes: Studies with Glutamate Dehydrogenase.
Mol.Pharmacol., 89, 2016
|
|
3JD4
 
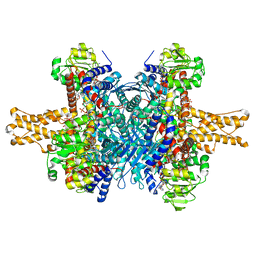 | | Glutamate dehydrogenase in complex with NADH and GTP, closed conformation | | 分子名称: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, GUANOSINE-5'-TRIPHOSPHATE, Glutamate dehydrogenase 1, ... | | 著者 | Borgnia, M.J, Banerjee, S, Merk, A, Matthies, D, Bartesaghi, A, Rao, P, Pierson, J, Earl, L.A, Falconieri, V, Subramaniam, S, Milne, J.L.S. | | 登録日 | 2016-03-28 | | 公開日 | 2016-04-27 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Using Cryo-EM to Map Small Ligands on Dynamic Metabolic Enzymes: Studies with Glutamate Dehydrogenase.
Mol.Pharmacol., 89, 2016
|
|
3JD1
 
 | | Glutamate dehydrogenase in complex with NADH, closed conformation | | 分子名称: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Glutamate dehydrogenase 1, mitochondrial | | 著者 | Borgnia, M.J, Banerjee, S, Merk, A, Matthies, D, Bartesaghi, A, Rao, P, Pierson, J, Earl, L.A, Falconieri, V, Subramaniam, S, Milne, J.L.S. | | 登録日 | 2016-03-28 | | 公開日 | 2016-04-27 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Using Cryo-EM to Map Small Ligands on Dynamic Metabolic Enzymes: Studies with Glutamate Dehydrogenase.
Mol.Pharmacol., 89, 2016
|
|
3JCZ
 
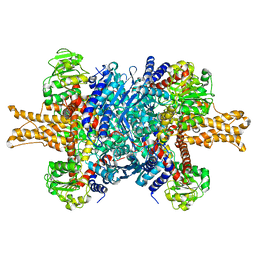 | | Structure of bovine glutamate dehydrogenase in the unliganded state | | 分子名称: | Glutamate dehydrogenase 1, mitochondrial | | 著者 | Borgnia, M.J, Banerjee, S, Merk, A, Matthies, D, Bartesaghi, A, Rao, P, Pierson, J, Earl, L.A, Falconieri, V, Subramaniam, S, Milne, J.L.S. | | 登録日 | 2016-03-27 | | 公開日 | 2016-04-27 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (3.26 Å) | | 主引用文献 | Using Cryo-EM to Map Small Ligands on Dynamic Metabolic Enzymes: Studies with Glutamate Dehydrogenase.
Mol.Pharmacol., 89, 2016
|
|
3JD3
 
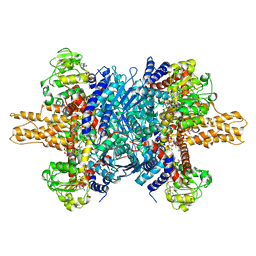 | | Glutamate dehydrogenase in complex with NADH and GTP, open conformation | | 分子名称: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, GUANOSINE-5'-TRIPHOSPHATE, Glutamate dehydrogenase 1, ... | | 著者 | Borgnia, M.J, Banerjee, S, Merk, A, Matthies, D, Bartesaghi, A, Rao, P, Pierson, J, Earl, L.A, Falconieri, V, Subramaniam, S, Milne, J.L.S. | | 登録日 | 2016-03-28 | | 公開日 | 2016-04-27 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Using Cryo-EM to Map Small Ligands on Dynamic Metabolic Enzymes: Studies with Glutamate Dehydrogenase.
Mol.Pharmacol., 89, 2016
|
|
3J7H
 
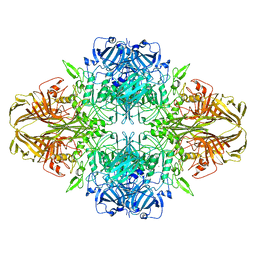 | | Structure of beta-galactosidase at 3.2-A resolution obtained by cryo-electron microscopy | | 分子名称: | Beta-galactosidase, MAGNESIUM ION | | 著者 | Bartesaghi, A, Matthies, D, Banerjee, S, Merk, A, Subramaniam, S. | | 登録日 | 2014-06-30 | | 公開日 | 2014-07-30 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structure of beta-galactosidase at 3.2- angstrom resolution obtained by cryo-electron microscopy.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3JD2
 
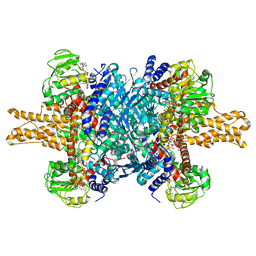 | | Glutamate dehydrogenase in complex with NADH, open conformation | | 分子名称: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Glutamate dehydrogenase 1, mitochondrial | | 著者 | Borgnia, M.J, Banerjee, S, Merk, A, Matthies, D, Bartesaghi, A, Rao, P, Pierson, J, Earl, L.A, Falconieri, V, Subramaniam, S, Milne, J.L.S. | | 登録日 | 2016-03-28 | | 公開日 | 2016-04-27 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Using Cryo-EM to Map Small Ligands on Dynamic Metabolic Enzymes: Studies with Glutamate Dehydrogenase.
Mol.Pharmacol., 89, 2016
|
|
8R47
 
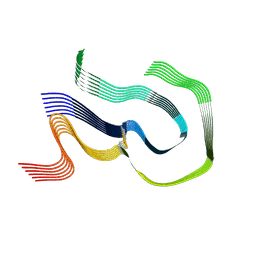 | | AL amyloid fibril from the FOR010 light chain | | 分子名称: | lambda 3 immunoglobulin light chain fragment, residues 4-116 | | 著者 | Pfeiffer, P.B, Banerjee, S, Schmidt, M, Faendrich, M. | | 登録日 | 2023-11-13 | | 公開日 | 2024-06-26 | | 実験手法 | ELECTRON MICROSCOPY (2.25 Å) | | 主引用文献 | Light chain mutations contribute to defining the fibril morphology in systemic AL amyloidosis.
Nat Commun, 15, 2024
|
|
8Q6S
 
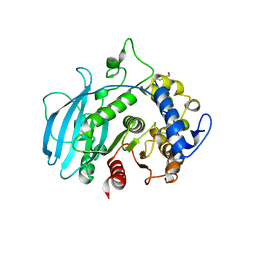 | | A carbohydrate esterase family 15 (CE15) glucuronoyl esterase from Phocaeicola vulgatus ATCC 8482 | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, Putative acetyl xylan esterase, ... | | 著者 | Mazurkewich, S, Seveso, A, Banerjee, S, Lo Leggio, L, Larsbrink, J. | | 登録日 | 2023-08-14 | | 公開日 | 2023-12-13 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.99 Å) | | 主引用文献 | Polysaccharide utilization loci from Bacteroidota encode CE15 enzymes with possible roles in cleaving pectin-lignin bonds.
Appl.Environ.Microbiol., 90, 2024
|
|
6N47
 
 | | The structure of SB-2-204-tubulin complex | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-(2-chloropyrido[3,2-d]pyrimidin-4-yl)-7-methoxy-3,4-dihydroquinoxalin-2(1H)-one, CALCIUM ION, ... | | 著者 | Arnst, K, Banerjee, S, Wang, Y, Li, W, Miller, D, Li, W. | | 登録日 | 2018-11-17 | | 公開日 | 2019-11-13 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | X-ray Crystal Structure Guided Discovery and Antitumor Efficacy of Dihydroquinoxalinone as Potent Tubulin Polymerization Inhibitors.
Acs Chem.Biol., 14, 2019
|
|
5K10
 
 | | Cryo-EM structure of isocitrate dehydrogenase (IDH1) | | 分子名称: | Isocitrate dehydrogenase [NADP] cytoplasmic, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Merk, A, Bartesaghi, A, Banerjee, S, Falconieri, V, Rao, P, Earl, L, Milne, J, Subramaniam, S. | | 登録日 | 2016-05-17 | | 公開日 | 2016-06-08 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Breaking Cryo-EM Resolution Barriers to Facilitate Drug Discovery.
Cell, 165, 2016
|
|
5K11
 
 | | Cryo-EM structure of isocitrate dehydrogenase (IDH1) in inhibitor-bound state | | 分子名称: | Isocitrate dehydrogenase [NADP] cytoplasmic, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Merk, A, Bartesaghi, A, Banerjee, S, Falconieri, V, Rao, P, Earl, L, Milne, J, Subramaniam, S. | | 登録日 | 2016-05-17 | | 公開日 | 2016-06-08 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Breaking Cryo-EM Resolution Barriers to Facilitate Drug Discovery.
Cell, 165, 2016
|
|
