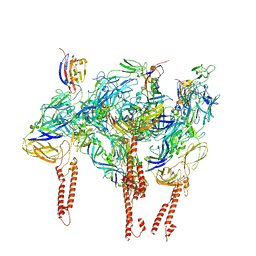
8DAN
 
 | |
8DAQ
 
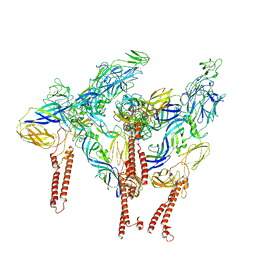 | |
8EWF
 
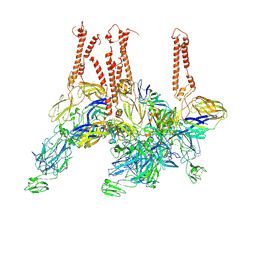 | |
8SQN
 
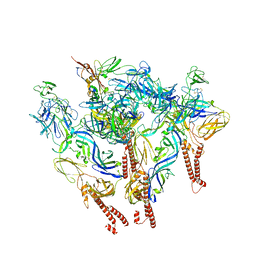 | | CryoEM structure of Western equine encephalitis virus VLP in complex with the chimeric Du-D1-Mo-D2 MXRA8 receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DuD1MoD2 chimeric MXRA8, E1 envelope glycoprotein, ... | | Authors: | Zimmerman, M.I, Fremont, D.H, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2023-05-04 | | Release date: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.89 Å) | | Cite: | Vertebrate-class-specific binding modes of the alphavirus receptor MXRA8.
Cell, 186, 2023
|
|
3CNV
 
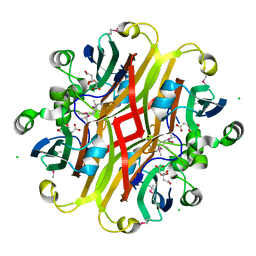 | | Crystal structure of the ligand-binding domain of a putative GntR-family transcriptional regulator from Bordetella bronchiseptica | | Descriptor: | CHLORIDE ION, CITRATE ANION, Putative GntR-family transcriptional regulator | | Authors: | Zimmerman, M.D, Xu, X, Cui, H, Filippova, E.V, Savchenko, A, Edwards, A.M, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-03-26 | | Release date: | 2008-04-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the ligand-binding domain of a putative GntR-family transcriptional regulator from Bordetella bronchiseptica.
To be Published
|
|
2FFS
 
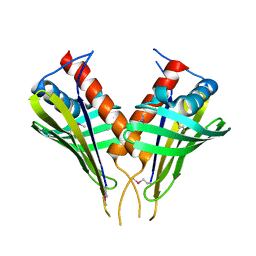 | | Structure of PR10-allergen-like protein PA1206 from Pseudomonas aeruginosa PAO1 | | Descriptor: | hypothetical protein PA1206 | | Authors: | Zimmerman, M.D, Chruszcz, M, Cymborowski, M.T, Wang, S, Kirillova, O, Skarina, T, Onopriyenko, O, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-12-20 | | Release date: | 2006-01-10 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of PR10-Allergen-Like Protein PA1206 From Pseudomonas aeruginosa PAO1
To be Published
|
|
2IAI
 
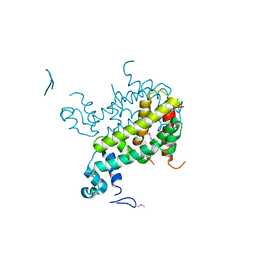 | | Crystal structure of SCO3833, a member of the TetR transcriptional regulator family from Streptomyces coelicolor A3 | | Descriptor: | Putative transcriptional regulator SCO3833 | | Authors: | Zimmerman, M.D, Xu, X, Wang, S, Gu, J, Chruszcz, M, Cymborowski, M, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-09-08 | | Release date: | 2006-09-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of SCO3833, a member of the TetR transcriptional regulator family from Streptomyces coelicolor A3
To be Published
|
|
2PAR
 
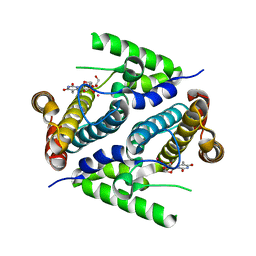 | | Crystal structure of the 5'-deoxynucleotidase YfbR mutant E72A complexed with Co(2+) and TMP | | Descriptor: | 5'-deoxynucleotidase YfbR, COBALT (II) ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Zimmerman, M.D, Proudfoot, M, Yakunin, A, Minor, W. | | Deposit date: | 2007-03-27 | | Release date: | 2008-03-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insight into the mechanism of substrate specificity and catalytic activity of an HD-domain phosphohydrolase: the 5'-deoxyribonucleotidase YfbR from Escherichia coli.
J.Mol.Biol., 378, 2008
|
|
2PAU
 
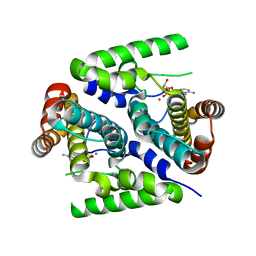 | | Crystal structure of the 5'-deoxynucleotidase YfbR mutant E72A complexed with Co(2+) and dAMP | | Descriptor: | 2'-DEOXYADENOSINE-5'-MONOPHOSPHATE, 5'-deoxynucleotidase YfbR, COBALT (II) ION, ... | | Authors: | Zimmerman, M.D, Proudfoot, M, Yakunin, A, Minor, W. | | Deposit date: | 2007-03-27 | | Release date: | 2008-03-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insight into the mechanism of substrate specificity and catalytic activity of an HD-domain phosphohydrolase: the 5'-deoxyribonucleotidase YfbR from Escherichia coli.
J.Mol.Biol., 378, 2008
|
|
2PAQ
 
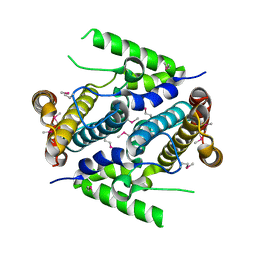 | | Crystal structure of the 5'-deoxynucleotidase YfbR | | Descriptor: | 5'-deoxynucleotidase YfbR | | Authors: | Zimmerman, M.D, Chruszcz, M, Cymborowski, M, Kudritska, M, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-03-27 | | Release date: | 2007-04-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insight into the mechanism of substrate specificity and catalytic activity of an HD-domain phosphohydrolase: the 5'-deoxyribonucleotidase YfbR from Escherichia coli.
J.Mol.Biol., 378, 2008
|
|
2KKH
 
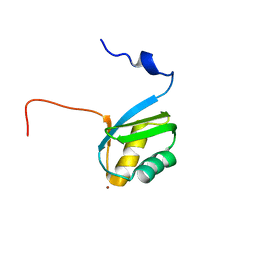 | | Structure of the zinc binding domain of the ATPase HMA4 | | Descriptor: | Putative heavy metal transporter, ZINC ION | | Authors: | Zimmerman, M, Clarke, O, Gulbis, J.M, Keizer, D.W, Jarvis, R.S, Cobbett, C.S, Hinds, M.G, Xiao, Z, Wedd, A.G. | | Deposit date: | 2009-06-20 | | Release date: | 2010-01-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Metal binding affinities of Arabidopsis zinc and copper transporters: selectivities match the relative, but not the absolute, affinities of their amino-terminal domains
Biochemistry, 48, 2009
|
|
3DXS
 
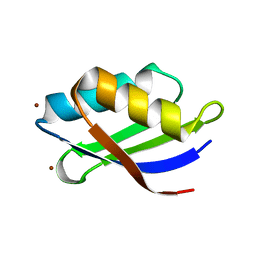 | | Crystal structure of a copper binding domain from HMA7, a P-type ATPase | | Descriptor: | Copper-transporting ATPase RAN1, LITHIUM ION, ZINC ION | | Authors: | Zimmermann, M, Xiao, Z, Clarke, O.B, Gulbis, J.M, Wedd, A.G. | | Deposit date: | 2008-07-25 | | Release date: | 2009-08-11 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Metal binding affinities of Arabidopsis zinc and copper transporters: selectivities match the relative, but not the absolute, affinities of their amino-terminal domains.
Biochemistry, 48, 2009
|
|
2M37
 
 | | Structure of lasso peptide astexin-1 | | Descriptor: | ASTEXIN-1 | | Authors: | Zimmermann, M, Hegemann, J.D, Xie, X, Marahiel, M.A. | | Deposit date: | 2013-01-14 | | Release date: | 2013-05-01 | | Method: | SOLUTION NMR | | Cite: | The astexin-1 lasso peptides: biosynthesis, stability, and structural studies.
Chem.Biol., 20, 2013
|
|
2MLJ
 
 | |
3K3S
 
 | | Crystal structure of altronate hydrolase (fragment 1-84) from Shigella Flexneri. | | Descriptor: | ACETATE ION, Altronate hydrolase, CHLORIDE ION, ... | | Authors: | Hou, J, Chruszcz, M, Xu, X, Le, B, Zimmerman, M.D, Savchenko, A, Edwards, A.M, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-10-04 | | Release date: | 2009-10-27 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of altronate hydrolase (fragment 1-84) from Shigella Flexneri.
To be Published
|
|
9BOG
 
 | | Structural basis for adhesin secretion by the outer-membrane usher in type 1 pili | | Descriptor: | Outer membrane usher protein FimD, Protein FimF, Type 1 fimbria chaperone FimC, ... | | Authors: | Bitter, R.M, Zimmerman, M, Hultgren, S, Yuan, P. | | Deposit date: | 2024-05-03 | | Release date: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.99 Å) | | Cite: | Structural basis for adhesin secretion by the outer-membrane usher in type 1 pili.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
5BQF
 
 | | Probable 2-hydroxyacid dehydrogenase from Rhizobium etli CFN 42 in complex with NADP, HEPES and L(+)-tartaric acid | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Langner, K.M, Shabalin, I.G, Handing, K.B, Gasiorowska, O.A, Stead, M, Hillerich, B.S, Chowdhury, S, Hammonds, J, Zimmerman, M.D, Al Obaidi, N, Bonanno, J, Seidel, R, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2015-05-29 | | Release date: | 2015-06-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Probable 2-hydroxyacid dehydrogenase from Rhizobium etli CFN 42 in complex with NADP, HEPES and L-tartaric acid
to be published
|
|
2FGC
 
 | | Crystal structure of Acetolactate synthase- small subunit from Thermotoga maritima | | Descriptor: | MAGNESIUM ION, acetolactate synthase, small subunit | | Authors: | Petkowski, J.J, Chruszcz, M, Zimmerman, M.D, Zheng, H, Cymborowski, M.T, Koclega, K.D, Kudritska, M, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-12-21 | | Release date: | 2006-02-07 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of TM0549 and NE1324--two orthologs of E. coli AHAS isozyme III small regulatory subunit.
Protein Sci., 16, 2007
|
|
4O9K
 
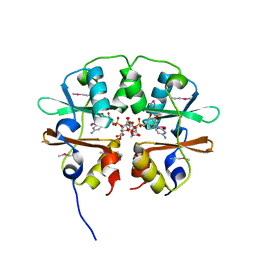 | | Crystal structure of the CBS pair of a putative D-arabinose 5-phosphate isomerase from Methylococcus capsulatus in complex with CMP-Kdo | | Descriptor: | Arabinose 5-phosphate isomerase, CYTIDINE 5'-MONOPHOSPHATE 3-DEOXY-BETA-D-GULO-OCT-2-ULO-PYRANOSONIC ACID, GLYCEROL | | Authors: | Shabalin, I.G, Cooper, D.R, Shumilin, I.A, Zimmerman, M.D, Majorek, K.A, Hammonds, J, Hillerich, B.S, Nawar, A, Bonanno, J, Seidel, R, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-01-02 | | Release date: | 2014-01-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure and kinetic properties of D-arabinose 5-phosphate isomerase from Methylococcus capsulatus
To be Published
|
|
4PMJ
 
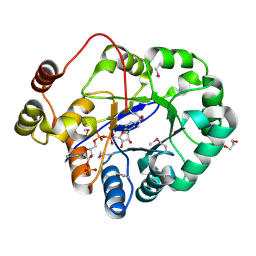 | | Crystal structure of a putative oxidoreductase from Sinorhizobium meliloti 1021 in complex with NADP | | Descriptor: | GLYCEROL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Putative oxidoreductase | | Authors: | Gasiorowska, O.A, Shabalin, I.G, Handing, K.B, Szlachta, K, Zimmerman, M.D, Hillerich, B.S, Gizzi, A, Toro, R, Bonanno, J, Seidel, R, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-05-21 | | Release date: | 2014-06-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a putative oxidoreductase from Sinorhizobiummeliloti 1021 in complex with NADP
to be published
|
|
3KZP
 
 | | Crystal structure of putative diguanylate cyclase/phosphodiesterase from Listaria monocytigenes | | Descriptor: | CACODYLATE ION, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Klimecka, M.M, Chruszcz, M, Zimmerman, M.D, Kudritska, M, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-12-08 | | Release date: | 2009-12-22 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of putative diguanylate cyclase/phosphodiesterase from Listaria monocytigenes
To be Published
|
|
3KXR
 
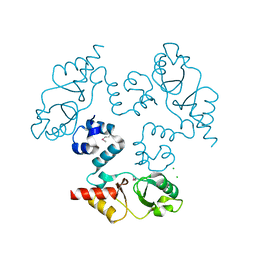 | | Structure of the cystathionine beta-synthase pair domain of the putative Mg2+ transporter SO5017 from Shewanella oneidensis MR-1. | | Descriptor: | CHLORIDE ION, Magnesium transporter, putative | | Authors: | Fratczak, Z, Zimmerman, M.D, Chruszcz, M, Cymborowski, M, Kagan, O, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-12-03 | | Release date: | 2009-12-15 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Structure of the cystathionine beta-synthase pair domain of the putative Mg2+ transporter SO5017 from Shewanella oneidensis MR-1.
To be Published
|
|
4JYJ
 
 | | Crystal Structure of putative enoyl-CoA hydratase/isomerase from Novosphingobium aromaticivorans DSM 12444 | | Descriptor: | Enoyl-CoA hydratase/isomerase, UNKNOWN LIGAND | | Authors: | Cooper, D.R, Porebski, P.J, Domagalski, M.J, Ahmed, M, Stead, M, Hillerich, B, Seidel, R, Zimmerman, M, Bonanno, J.B, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-03-29 | | Release date: | 2013-05-29 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of putative enoyl-CoA hydratase/isomerase from Novosphingobium aromaticivorans DSM 12444
To be Published
|
|
4JVT
 
 | | Crystal structure of Tfu_1878, a putative enoyl-CoA hydratase fromThermobifida fusca YX in complex with CoA | | Descriptor: | ACETATE ION, ACETYL COENZYME *A, Enoyl-CoA hydratase | | Authors: | Mikolajczak, K, Porebski, P.J, Cooper, D.R, Ahmed, M, Stead, M, Hillerich, B, Seidel, R, Zimmerman, M, Bonanno, J.B, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-03-26 | | Release date: | 2013-06-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of Tfu_1878, a putative enoyl-CoA hydratase fromThermobifida fusca YX in complex with CoA
To be Published
|
|
3N73
 
 | | Crystal structure of a putative 4-hydroxy-2-oxoglutarate aldolase from Bacillus cereus | | Descriptor: | CHLORIDE ION, Putative 4-hydroxy-2-oxoglutarate aldolase | | Authors: | Cabello, R, Chruszcz, M, Xu, X, Zimmerman, M.D, Cui, H, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-05-26 | | Release date: | 2010-06-09 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Crystal structure of a putative 4-hydroxy-2-oxoglutarate aldolase from Bacillus cereus
To be Published
|
|
