7WM0
 
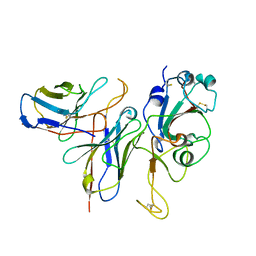 | |
5ZHU
 
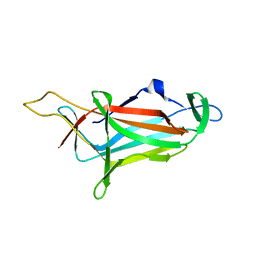 | |
8IVP
 
 | | Crystal structure of MV in complex with LLP and FRU from Mycobacterium vanbaalenii | | Descriptor: | Branched chain amino acid: 2-keto-4-methylthiobutyrate aminotransferase, D-fructose | | Authors: | Li, Q, Zhu, Y.M, Gao, J, Wei, H.L, Han, X, Liu, W.D, Sun, Y.X. | | Deposit date: | 2023-03-28 | | Release date: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structure of MV in complex with LLP and FRU from Mycobacterium vanbaalenii
To Be Published
|
|
4EF4
 
 | | Crystal structure of STING CTD complex with c-di-GMP | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), CALCIUM ION, Transmembrane protein 173 | | Authors: | Ouyang, S, Ru, H, Shaw, N, Jiang, Y, Niu, F, Zhu, Y, Qiu, W, Li, Y, Liu, Z.-J. | | Deposit date: | 2012-03-29 | | Release date: | 2012-05-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.147 Å) | | Cite: | Structural analysis of the STING adaptor protein reveals a hydrophobic dimer interface and mode of cyclic di-GMP binding
Immunity, 36, 2012
|
|
4EAC
 
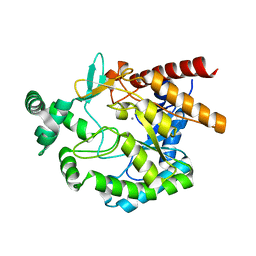 | | Crystal structure of mannonate dehydratase from Escherichia coli strain K12 | | Descriptor: | CHLORIDE ION, MANGANESE (II) ION, Mannonate dehydratase | | Authors: | Qiu, X, Zhu, Y, Yuan, Y, Zhang, Y, Liu, H, Gao, Y, Teng, M, Niu, L. | | Deposit date: | 2012-03-22 | | Release date: | 2013-03-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insights into decreased enzymatic activity induced by an insert sequence in mannonate dehydratase from Gram negative bacterium.
J.Struct.Biol., 180, 2012
|
|
5XBL
 
 | | Structure of nuclease in complex with associated protein | | Descriptor: | Associated protein, CRISPR-associated endonuclease Cas9/Csn1, RNA (98-MER) | | Authors: | Dong, D, Guo, M, Wang, S, Zhu, Y, Huang, Z. | | Deposit date: | 2017-03-20 | | Release date: | 2017-06-14 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.052 Å) | | Cite: | Structural basis of CRISPR-SpyCas9 inhibition by an anti-CRISPR protein
Nature, 546, 2017
|
|
7VCI
 
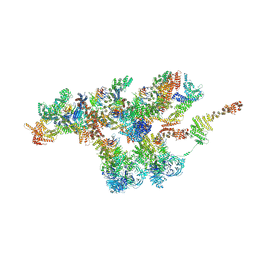 | | Structure of Xenopus laevis NPC nuclear ring asymmetric unit | | Descriptor: | GATOR complex protein SEC13, MGC154553 protein, MGC83295 protein, ... | | Authors: | Tai, L, Zhu, Y, Sun, F. | | Deposit date: | 2021-09-03 | | Release date: | 2022-02-02 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (8.1 Å) | | Cite: | 8 angstrom structure of the outer rings of the Xenopus laevis nuclear pore complex obtained by cryo-EM and AI.
Protein Cell, 13, 2022
|
|
6JJ4
 
 | | Crystal structure of OsHXK6-apo form | | Descriptor: | Hexokinase-6 | | Authors: | He, C, Wei, P, Chen, J, Wang, H, Wan, Y, Zhou, J, Zhu, Y, Huang, W, Yin, L. | | Deposit date: | 2019-02-25 | | Release date: | 2019-07-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of OsHXK6-apo
To Be Published
|
|
6JJ9
 
 | | Crystal structure of OsHXK6-Glc-ATP-Mg2+ complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Hexokinase-6, MAGNESIUM ION, ... | | Authors: | He, C, Wei, P, Chen, J, Wang, H, Wan, Y, Zhou, J, Zhu, Y, Huang, W, Yin, L. | | Deposit date: | 2019-02-25 | | Release date: | 2019-07-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of OsHXK6-Glc-ATP-Mg2+ complex
To Be Published
|
|
6K2U
 
 | | Crystal structure of Thr66 ADP-ribosylated ubiquitin | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, MAGNESIUM ION, Polyubiquitin-C, ... | | Authors: | Wang, X, Zhou, Y, Zhu, Y. | | Deposit date: | 2019-05-15 | | Release date: | 2020-03-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.554 Å) | | Cite: | Threonine ADP-Ribosylation of Ubiquitin by a Bacterial Effector Family Blocks Host Ubiquitination.
Mol.Cell, 78, 2020
|
|
7WLZ
 
 | |
7WLY
 
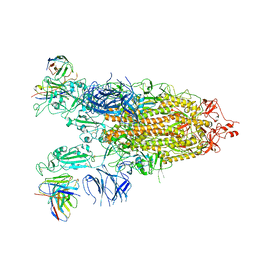 | | Cryo-EM structure of the Omicron S in complex with 35B5 Fab(1 down- and 2 up RBDs) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of 35B5 Fab, ... | | Authors: | Wang, X, Zhu, Y. | | Deposit date: | 2022-01-14 | | Release date: | 2022-05-25 | | Last modified: | 2022-06-22 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | 35B5 antibody potently neutralizes SARS-CoV-2 Omicron by disrupting the N-glycan switch via a conserved spike epitope.
Cell Host Microbe, 30, 2022
|
|
6JJ8
 
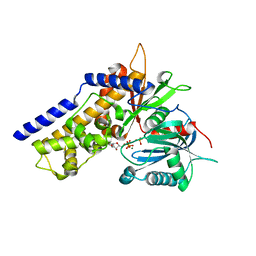 | | Crystal structure of OsHXK6-ATP-Mg2+ complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | He, C, Wei, P, Chen, J, Wang, H, Wan, Y, Zhou, J, Zhu, Y, Huang, W, Yin, L. | | Deposit date: | 2019-02-25 | | Release date: | 2019-07-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of OsHXK6-ATP-Mg2+ complex
To Be Published
|
|
5X6D
 
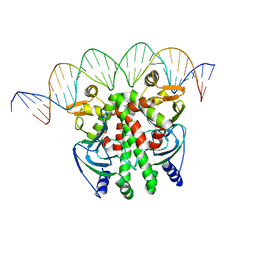 | | Crystal structure of PrfA-DNA binary complex | | Descriptor: | DNA (28-MER), DNA (29-MER), Listeriolysin positive regulatory factor A | | Authors: | Wang, Y, Feng, H, Zhu, Y.L, Gao, P. | | Deposit date: | 2017-02-21 | | Release date: | 2017-04-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Structural insights into glutathione-mediated activation of the master regulator PrfA in Listeria monocytogenes
Protein Cell, 8, 2017
|
|
7WSO
 
 | | Structure of a membrane protein G | | Descriptor: | B-cell antigen receptor complex-associated protein alpha chain, B-cell antigen receptor complex-associated protein beta chain, Immunoglobulin heavy constant gamma 1 | | Authors: | Ma, X, Zhu, Y, Chen, Y, Huang, Z. | | Deposit date: | 2022-01-30 | | Release date: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Cryo-EM structures of two human B cell receptor isotypes.
Science, 377, 2022
|
|
5X6E
 
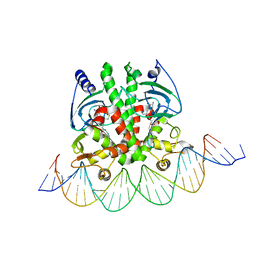 | | Crystal structure of PrfA-DNA binary complex | | Descriptor: | DNA (28-MER), DNA (29-MER), GLUTATHIONE, ... | | Authors: | Wang, Y, Feng, H, Zhu, Y.L, Gao, P. | | Deposit date: | 2017-02-21 | | Release date: | 2017-04-12 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Structural insights into glutathione-mediated activation of the master regulator PrfA in Listeria monocytogenes
Protein Cell, 8, 2017
|
|
6JJ7
 
 | | Crystal structure of OsHXK6-Glc complex | | Descriptor: | Rice hexokinase 6, beta-D-glucopyranose | | Authors: | He, C, Wei, P, Chen, J, Wang, H, Wan, Y, Zhou, J, Zhu, Y, Huang, W, Yin, L. | | Deposit date: | 2019-02-25 | | Release date: | 2019-07-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of OsHXK6-Glc complex
To Be Published
|
|
3VNI
 
 | | Crystal structures of D-Psicose 3-epimerase from Clostridium cellulolyticum H10 and its complex with ketohexose sugars | | Descriptor: | MANGANESE (II) ION, Xylose isomerase domain protein TIM barrel | | Authors: | Chan, H.C, Zhu, Y, Hu, Y, Ko, T.P, Huang, C.H, Ren, F, Chen, C.C, Guo, R.T, Sun, Y. | | Deposit date: | 2012-01-16 | | Release date: | 2012-08-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystal structures of D-psicose 3-epimerase from Clostridium cellulolyticum H10 and its complex with ketohexose sugars.
Protein Cell, 3, 2012
|
|
7X8X
 
 | | structural insights into Mycobacterium tuberculosis ClpP1P2 inhibition by Cediranib: implications for developing antimicrobial agents targeting Clp protease | | Descriptor: | 4-[1-[3-[4-[(4-fluoranyl-2-methyl-1H-indol-5-yl)oxy]-6-methoxy-quinazolin-7-yl]oxypropyl]piperidin-4-yl]benzamide, 4-[[3,5-bis(fluoranyl)phenyl]methyl]-N-[(4-bromophenyl)methyl]piperazine-1-carboxamide, ATP-dependent Clp protease proteolytic subunit 1, ... | | Authors: | Bao, R, Luo, Y.F, Zhu, Y.B, Yang, Y, Zhou, Y.Z. | | Deposit date: | 2022-03-15 | | Release date: | 2023-04-12 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (3.24 Å) | | Cite: | Discovery and Mechanistic Study of Novel Mycobacterium tuberculosis ClpP1P2 Inhibitors
J.Med.Chem., 66, 2023
|
|
3VNJ
 
 | | Crystal structures of D-Psicose 3-epimerase with D-psicose from Clostridium cellulolyticum H10 | | Descriptor: | D-psicose, MANGANESE (II) ION, Xylose isomerase domain protein TIM barrel | | Authors: | Chan, H.C, Zhu, Y, Hu, Y, Ko, T.P, Huang, C.H, Ren, F, Chen, C.C, Guo, R.T, Sun, Y. | | Deposit date: | 2012-01-16 | | Release date: | 2012-08-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Crystal structures of D-psicose 3-epimerase from Clostridium cellulolyticum H10 and its complex with ketohexose sugars.
Protein Cell, 3, 2012
|
|
3VNM
 
 | | Crystal structures of D-Psicose 3-epimerase with D-sorbose from Clostridium cellulolyticum H10 | | Descriptor: | D-sorbose, MANGANESE (II) ION, Xylose isomerase domain protein TIM barrel | | Authors: | Chan, H.C, Zhu, Y, Hu, Y, Ko, T.P, Huang, C.H, Ren, F, Chen, C.C, Guo, R.T, Sun, Y. | | Deposit date: | 2012-01-17 | | Release date: | 2012-08-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Crystal structures of D-psicose 3-epimerase from Clostridium cellulolyticum H10 and its complex with ketohexose sugars.
Protein Cell, 3, 2012
|
|
3VNK
 
 | | Crystal structures of D-Psicose 3-epimerase with D-fructose from Clostridium cellulolyticum H10 | | Descriptor: | D-fructose, MANGANESE (II) ION, Xylose isomerase domain protein TIM barrel | | Authors: | Chan, H.C, Zhu, Y, Hu, Y, Ko, T.P, Huang, C.H, Ren, F, Chen, C.C, Guo, R.T, Sun, Y. | | Deposit date: | 2012-01-16 | | Release date: | 2012-08-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Crystal structures of D-psicose 3-epimerase from Clostridium cellulolyticum H10 and its complex with ketohexose sugars.
Protein Cell, 3, 2012
|
|
3VNL
 
 | | Crystal structures of D-Psicose 3-epimerase with D-tagatose from Clostridium cellulolyticum H10 | | Descriptor: | D-tagatose, MANGANESE (II) ION, Xylose isomerase domain protein TIM barrel | | Authors: | Chan, H.C, Zhu, Y, Hu, Y, Ko, T.P, Huang, C.H, Ren, F, Chen, C.C, Guo, R.T, Sun, Y. | | Deposit date: | 2012-01-16 | | Release date: | 2012-08-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structures of D-psicose 3-epimerase from Clostridium cellulolyticum H10 and its complex with ketohexose sugars.
Protein Cell, 3, 2012
|
|
8H77
 
 | | Hsp90-AhR-p23-XAP2 complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, AH receptor-interacting protein, Aryl hydrocarbon receptor, ... | | Authors: | Wen, Z.L, Zhai, Y.J, Zhu, Y, Sun, F. | | Deposit date: | 2022-10-19 | | Release date: | 2023-01-04 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure of the cytosolic AhR complex.
Structure, 31, 2023
|
|
7EL1
 
 | | Structure of a protein from bacteria | | Descriptor: | 100AA, CRISPR-associated endonuclease Cas9, DNA (28-MER), ... | | Authors: | Liu, H, Zhu, Y, Huang, Z. | | Deposit date: | 2021-04-07 | | Release date: | 2021-07-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structural basis of Staphylococcus aureus Cas9 inhibition by AcrIIA14.
Nucleic Acids Res., 49, 2021
|
|
