6IV6
 
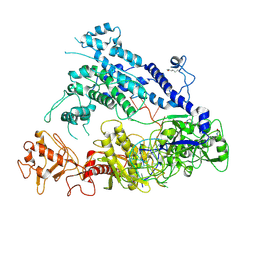 | | Cryo-EM structure of AcrVA5-acetylated MbCas12a in complex with crRNA | | Descriptor: | RNA (59-MER), nuclease | | Authors: | Dong, L, Li, N, Guan, X, Zhu, Y, Gao, N, Huang, Z. | | Deposit date: | 2018-12-02 | | Release date: | 2019-04-10 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | An anti-CRISPR protein disables type V Cas12a by acetylation.
Nat. Struct. Mol. Biol., 26, 2019
|
|
8WHT
 
 | |
8WLI
 
 | |
8WK3
 
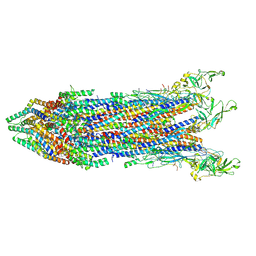 | | Cryo-EM structure of the proximal rod-export apparatus and FlgF within the motor-hook complex in the CW state | | Descriptor: | Flagellar M-ring protein, Flagellar basal body rod protein FlgB, Flagellar basal-body rod protein FlgC, ... | | Authors: | Tan, J.X, Zhang, L, Zhou, Y, Zhu, Y.Q. | | Deposit date: | 2023-09-26 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structure of the proximal rod-export apparatus and FlgF within the motor-hook complex in the CW state
To Be Published
|
|
8WLE
 
 | |
8WJR
 
 | |
8WL2
 
 | | Cryo-EM structure of the membrane-anchored part of the flagellar motor-hook complex in the CW state. | | Descriptor: | Flagellar L-ring protein, Flagellar M-ring protein, Flagellar P-ring protein, ... | | Authors: | Tan, J.X, Zhang, L, Zhou, Y, Zhu, Y.Q. | | Deposit date: | 2023-09-29 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structure of the membrane-anchored part of the flagellar motor-hook complex in the CW state.
To Be Published
|
|
8WLP
 
 | |
8WLH
 
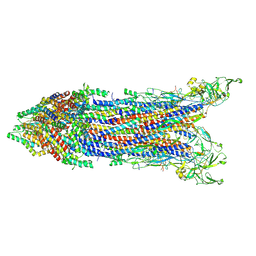 | | Cryo-EM structure of the proximal rod-export apparatus and FlgF within the motor-hook complex in the CCW state | | Descriptor: | Flagellar M-ring protein, Flagellar basal body rod protein FlgB, Flagellar basal-body rod protein FlgC, ... | | Authors: | Tan, J.X, Zhang, L, Zhou, Y, Zhu, Y.Q. | | Deposit date: | 2023-09-29 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structure of the proximal rod-export apparatus and FlgF within the motor-hook complex in the CCW state
To Be Published
|
|
8WO5
 
 | | Cryo-EM structure of the intact flagellar motor-hook complex in the CCW state | | Descriptor: | Flagellar L-ring protein, Flagellar M-ring protein, Flagellar P-ring protein, ... | | Authors: | Tan, J.X, Zhang, L, Zhou, Y, Zhu, Y.Q. | | Deposit date: | 2023-10-06 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (7.4 Å) | | Cite: | Cryo-EM structure of the intact flagellar motor-hook complex in the CCW state
To Be Published
|
|
8WK4
 
 | |
8WKI
 
 | |
8XP0
 
 | | Cryo-EM structure of the protomers of the C ring in the CCW state | | Descriptor: | Flagellar M-ring protein, Flagellar motor switch protein FliG, Flagellar motor switch protein FliM, ... | | Authors: | Tan, J.X, Zhang, L, Zhou, Y, Zhu, Y.Q. | | Deposit date: | 2024-01-02 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM structure of the protomers of the C ring in the CCW state
To Be Published
|
|
4M36
 
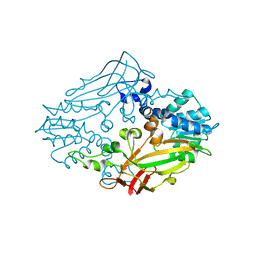 | |
4M37
 
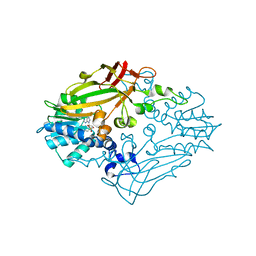 | |
4NFZ
 
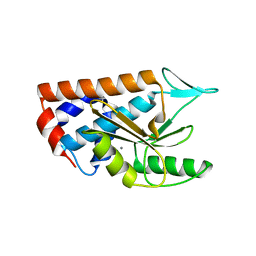 | | Crystal structure of polymerase subunit PA N-terminal endonuclease domain from bat-derived influenza virus H17N10 | | Descriptor: | MANGANESE (II) ION, Polymerase PA | | Authors: | Tefsen, B, Lu, G, Zhu, Y, Haywood, J, Zhao, L, Deng, T, Qi, J, Gao, G.F. | | Deposit date: | 2013-11-01 | | Release date: | 2013-12-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The N-Terminal Domain of PA from Bat-Derived Influenza-Like Virus H17N10 Has Endonuclease Activity
J.Virol., 88, 2014
|
|
8WLQ
 
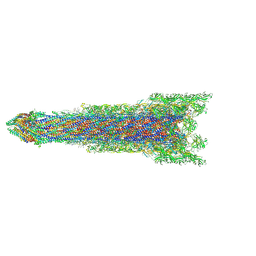 | | Cryo-EM structure of the whole rod-export apparatus with hook within the flagellar motor-hook complex in the CCW state. | | Descriptor: | Flagellar M-ring protein, Flagellar basal body rod protein FlgB, Flagellar basal-body rod protein FlgC, ... | | Authors: | Tan, J.X, Zhang, L, Zhou, Y, Zhu, Y.Q. | | Deposit date: | 2023-09-30 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structure of the whole rod-export apparatus with hook within the flagellar motor-hook complex in the CCW state.
To Be Published
|
|
8WLT
 
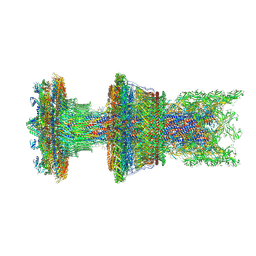 | | Cryo-EM structure of the membrane-anchored part of the flagellar motor-hook complex in the CCW state | | Descriptor: | Flagellar L-ring protein, Flagellar M-ring protein, Flagellar P-ring protein, ... | | Authors: | Tan, J.X, Zhang, L, Zhou, Y, Zhu, Y.Q. | | Deposit date: | 2023-10-01 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM structure of the membrane-anchored part of the flagellar motor-hook complex in the CCW state
To Be Published
|
|
8WKQ
 
 | | Cryo-EM structure of the MS ring (C1) with export apparatus and proximal rod within the flagellar motor-hook complex in the CW state. | | Descriptor: | Flagellar M-ring protein, Flagellar basal body rod protein FlgB, Flagellar basal-body rod protein FlgC, ... | | Authors: | Tan, J.X, Zhang, L, Zhou, Y, Zhu, Y.Q. | | Deposit date: | 2023-09-28 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structure of the MS ring (C1) with export apparatus and proximal rod within the flagellar motor-hook complex in the CW state.
To Be Published
|
|
8WKK
 
 | | Cryo-EM structure of the whole rod with export apparatus and hook within the flagellar motor-hook complex in the CW state. | | Descriptor: | Flagellar M-ring protein, Flagellar basal body rod protein FlgB, Flagellar basal-body rod protein FlgC, ... | | Authors: | Tan, J.X, Zhang, L, Zhou, Y, Zhu, Y.Q. | | Deposit date: | 2023-09-28 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structure of the whole rod with export apparatus and hook within the flagellar motor-hook complex in the CW state.
To Be Published
|
|
8WOE
 
 | | Cryo-EM structure of the intact flagellar motor-hook complex in the CW state | | Descriptor: | Chemotaxis protein CheY, Flagellar L-ring protein, Flagellar M-ring protein, ... | | Authors: | Tan, J.X, Zhang, L, Zhou, Y, Zhu, Y.Q. | | Deposit date: | 2023-10-07 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Cryo-EM structure of the intact flagellar motor-hook complex in the CW state
To Be Published
|
|
8WLN
 
 | | Cryo-EM structure of the MS ring with export apparatus and proximal rod within the motor-hook complex in the CCW state | | Descriptor: | Flagellar M-ring protein, Flagellar basal body rod protein FlgB, Flagellar basal-body rod protein FlgC, ... | | Authors: | Tan, J.X, Zhang, L, Zhou, Y, Zhu, Y.Q. | | Deposit date: | 2023-09-30 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Cryo-EM structure of the MS ring with export apparatus and proximal rod within the motor-hook complex in the CCW state
To Be Published
|
|
8WIW
 
 | | Cryo-EM structure of the flagellar C ring in the CW state | | Descriptor: | Chemotaxis protein CheY, Flagellar M-ring protein, Flagellar motor switch protein FliG, ... | | Authors: | Tan, J.X, Zhang, L, Zhou, Y, Zhu, Y.Q. | | Deposit date: | 2023-09-25 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (5.6 Å) | | Cite: | Cryo-EM structure of the flagellar C ring in the CW state
To Be Published
|
|
7Y04
 
 | | Hsp90-AhR-p23 complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Aryl hydrocarbon receptor, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Wen, Z.L, Zhai, Y.J, Zhu, Y, Sun, F. | | Deposit date: | 2022-06-03 | | Release date: | 2023-01-04 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structure of the cytosolic AhR complex.
Structure, 31, 2023
|
|
8IOZ
 
 | | Crystal structure of transaminase | | Descriptor: | Branched chain amino acid: 2-keto-4-methylthiobutyrate aminotransferase | | Authors: | Li, Q, Zhu, Y.M, Gao, J, Wei, H.L, Han, X, Liu, W.D, Sun, Y.X. | | Deposit date: | 2023-03-13 | | Release date: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | structure of aminotransferase
To Be Published
|
|
