5TI2
 
 | | CRYSTAL STRUCTURE OF THE FIRST BROMODOMAIN OF HUMAN BRD4 IN COMPLEX WITH INHIBITOR 7635936 | | Descriptor: | 1,2-ETHANEDIOL, 3-fluoro-N-[3-(2-oxopyrrolidin-1-yl)phenyl]benzene-1-sulfonamide, Bromodomain-containing protein 4 | | Authors: | Zhu, J.-Y, Ember, S.W.J, SCHONBRUNN, E. | | Deposit date: | 2016-09-30 | | Release date: | 2017-08-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Identification of a Novel Class of BRD4 Inhibitors by Computational Screening and Binding Simulations.
ACS Omega, 2, 2017
|
|
5TI7
 
 | | CRYSTAL STRUCTURE OF THE FIRST BROMODOMAIN OF HUMAN BRD4 IN COMPLEX WITH INHIBITOR 17528462 | | Descriptor: | 1,2-ETHANEDIOL, 5-bromo-2-methoxy-N-(3-(2-oxopyrrolidin-1-yl)phenyl)benzenesulfonamide, Bromodomain-containing protein 4 | | Authors: | Zhu, J.-Y, Ember, S.W.J, Schonbrunn, E. | | Deposit date: | 2016-10-01 | | Release date: | 2017-08-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Identification of a Novel Class of BRD4 Inhibitors by Computational Screening and Binding Simulations.
ACS Omega, 2, 2017
|
|
5TI6
 
 | | CRYSTAL STRUCTURE OF THE FIRST BROMODOMAIN OF HUMAN BRD4 IN COMPLEX WITH INHIBITOR 8841881 | | Descriptor: | 1,2-ETHANEDIOL, 2,6-difluoro-N-[3-(2-oxopyrrolidin-1-yl)phenyl]benzene-1-sulfonamide, Bromodomain-containing protein 4 | | Authors: | Zhu, J.-Y, Ember, S.W.J, Schonbrunn, E. | | Deposit date: | 2016-10-01 | | Release date: | 2017-08-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.704 Å) | | Cite: | Identification of a Novel Class of BRD4 Inhibitors by Computational Screening and Binding Simulations.
ACS Omega, 2, 2017
|
|
5TI5
 
 | | CRYSTAL STRUCTURE OF THE FIRST BROMODOMAIN OF HUMAN BRD4 IN COMPLEX WITH INHIBITOR 8841880 | | Descriptor: | 1,2-ETHANEDIOL, 3-bromo-N-[3-(2-oxo-2,3-dihydro-1H-pyrrol-1-yl)phenyl]benzene-1-sulfonamide, Bromodomain-containing protein 4 | | Authors: | Zhu, J.-Y, Ember, S.W.J, Schonbrunn, E. | | Deposit date: | 2016-10-01 | | Release date: | 2017-08-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Identification of a Novel Class of BRD4 Inhibitors by Computational Screening and Binding Simulations.
ACS Omega, 2, 2017
|
|
5TI4
 
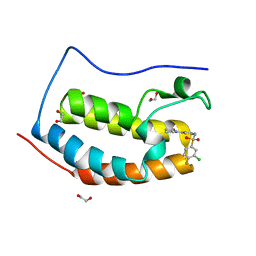 | | CRYSTAL STRUCTURE OF THE FIRST BROMODOMAIN OF HUMAN BRD4 IN COMPLEX WITH INHIBITOR 8841871 | | Descriptor: | 1,2-ETHANEDIOL, 3-chloro-4-fluoro-N-[3-(2-oxopyrrolidin-1-yl)phenyl]benzene-1-sulfonamide, Bromodomain-containing protein 4 | | Authors: | Zhu, J.-Y, Ember, S.W.J, SCHONBRUNN, E. | | Deposit date: | 2016-09-30 | | Release date: | 2017-08-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Identification of a Novel Class of BRD4 Inhibitors by Computational Screening and Binding Simulations.
ACS Omega, 2, 2017
|
|
6KKG
 
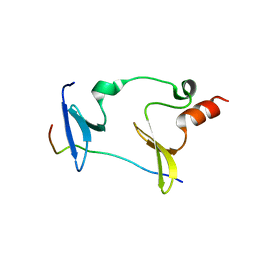 | | Crystal structure of MAGI2-Dendrin complex | | Descriptor: | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 2, Peptide from Dendrin | | Authors: | Zhu, J.W, Zhang, H.J, Lin, L, Zhang, R.G. | | Deposit date: | 2019-07-25 | | Release date: | 2020-07-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Phase separation of MAGI2-mediated complex underlies formation of slit diaphragm complex in glomerular filtration barrier
To Be Published
|
|
6JMT
 
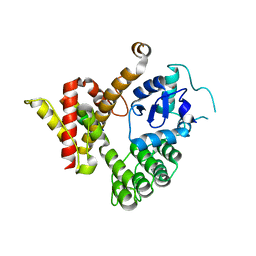 | | Crystal structure of GIT/PIX complex | | Descriptor: | ARF GTPase-activating protein GIT2, ZINC ION, beta PIX | | Authors: | Zhu, J, Lin, L, Xia, Y, Zhang, R, Zhang, M. | | Deposit date: | 2019-03-13 | | Release date: | 2020-05-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | GIT/PIX Condensates Are Modular and Ideal for Distinct Compartmentalized Cell Signaling.
Mol.Cell, 79, 2020
|
|
7VS4
 
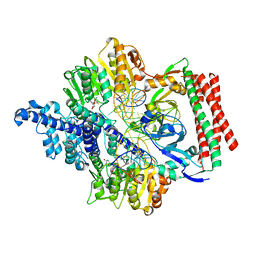 | | Crystal structure of PacII_M1M2S-DNA(m6A)-SAH complex | | Descriptor: | DNA (25-mer), S-ADENOSYL-L-HOMOCYSTEINE, Site-specific DNA recognition subunit, ... | | Authors: | Zhu, J, Gao, P. | | Deposit date: | 2021-10-25 | | Release date: | 2022-11-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Molecular insights into DNA recognition and methylation by non-canonical type I restriction-modification systems.
Nat Commun, 13, 2022
|
|
7VRU
 
 | | Crystal structure of PacII_M1M2S-DNA-SAH complex | | Descriptor: | DNA (25-mer), S-ADENOSYL-L-HOMOCYSTEINE, Site-specific DNA recognition subunit, ... | | Authors: | Zhu, J, Gao, P. | | Deposit date: | 2021-10-25 | | Release date: | 2022-11-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular insights into DNA recognition and methylation by non-canonical type I restriction-modification systems.
Nat Commun, 13, 2022
|
|
6JMU
 
 | | Crystal structure of GIT1/Paxillin complex | | Descriptor: | ARF GTPase-activating protein GIT1, Paxillin | | Authors: | Zhu, J, Lin, L, Xia, Y, Zhang, R, Zhang, M. | | Deposit date: | 2019-03-13 | | Release date: | 2020-05-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | GIT/PIX Condensates Are Modular and Ideal for Distinct Compartmentalized Cell Signaling.
Mol.Cell, 79, 2020
|
|
7B57
 
 | | Crystal structure of CaMKII-actinin complex bound to ADP | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ADENOSINE-5'-DIPHOSPHATE, Alpha-actinin-2, ... | | Authors: | Zhu, J, Gold, M. | | Deposit date: | 2020-12-03 | | Release date: | 2022-06-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of CaMKII-actinin complex bound to ADP
To Be Published
|
|
7B55
 
 | | Crystal structure of CaMKII-actinin complex bound to MES | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Alpha-actinin-2, Calcium/calmodulin-dependent protein kinase type II subunit alpha | | Authors: | Zhu, J, Gold, M. | | Deposit date: | 2020-12-03 | | Release date: | 2022-06-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of CaMKII-actinin complex bound to MES
To Be Published
|
|
7B56
 
 | | Crystal structure of CaMKII-actinin complex bound to AMPPNP | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Alpha-actinin-2, Calcium/calmodulin-dependent protein kinase type II subunit alpha, ... | | Authors: | Zhu, J, Gold, M. | | Deposit date: | 2020-12-03 | | Release date: | 2022-06-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure of CaMKII-actinin complex bound to MES
To Be Published
|
|
6QST
 
 | | Structure of CREBBP bromodomain with compound 2 bound | | Descriptor: | CREB-binding protein, ~{N}-[3-(3-azanyl-5-methyl-1,2-oxazol-4-yl)-5-(5-ethanoyl-2-ethoxy-phenyl)phenyl]furan-2-carboxamide | | Authors: | Zhu, J, Sledz, P, Caflisch, A. | | Deposit date: | 2019-02-22 | | Release date: | 2020-03-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of CREBBP bromodomain with compound 2 bound
To Be Published
|
|
5VD7
 
 | | CRYSTAL STRUCTURE OF HUMAN WEE1 KINASE DOMAIN IN COMPLEX WITH RAC-IV-098, a MK1775 analogue | | Descriptor: | 1,2-ETHANEDIOL, 6-{[3-fluoro-4-(4-methylpiperazin-1-yl)phenyl]amino}-1-[6-(2-hydroxypropan-2-yl)pyridin-2-yl]-2-(prop-2-en-1-yl)-1,2-dihydro-3H-pyrazolo[3,4-d]pyrimidin-3-one, CHLORIDE ION, ... | | Authors: | Zhu, J.-Y, Schonbrunn, E. | | Deposit date: | 2017-04-01 | | Release date: | 2018-04-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structural basis of Wee family kinase inhibition by small molecules
to be published
|
|
5VD5
 
 | |
5VDA
 
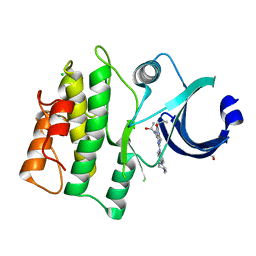 | | Crystal structure of human WEE1 kinase domain in complex with RAC-IV-101, a MK1775 analogue | | Descriptor: | 1,2-ETHANEDIOL, 1-{6-[(1S)-1-hydroxyethyl]pyridin-2-yl}-6-{[4-(4-methylpiperazin-1-yl)phenyl]amino}-2-(prop-2-en-1-yl)-1,2-dihydro-3H-pyrazolo[3,4-d]pyrimidin-3-one, CHLORIDE ION, ... | | Authors: | Zhu, J.-Y, Schonbrunn, E. | | Deposit date: | 2017-04-01 | | Release date: | 2018-04-04 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of Wee family kinase inhibition by small molecules
to be published
|
|
5VD9
 
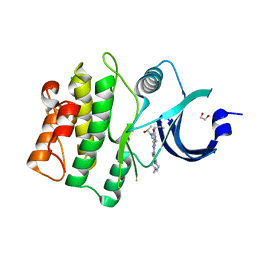 | | Crystal structure of human WEE1 kinase domain in complex with RAC-IV-097, a MK1775 analogue | | Descriptor: | 1,2-ETHANEDIOL, 1-{6-[(1R)-1-hydroxyethyl]pyridin-2-yl}-6-{[4-(4-methylpiperazin-1-yl)phenyl]amino}-2-(prop-2-en-1-yl)-1,2-dihydro-3H-pyrazolo[3,4-d]pyrimidin-3-one, CHLORIDE ION, ... | | Authors: | Zhu, J.-Y, Schonbrunn, E. | | Deposit date: | 2017-04-01 | | Release date: | 2018-04-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural basis of Wee family kinase inhibition by small molecules
to be published
|
|
5VD4
 
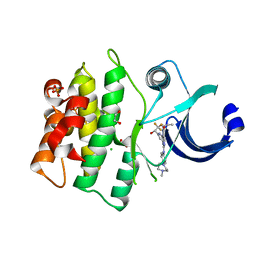 | | CRYSTAL STRUCTURE OF HUMAN WEE1 KINASE DOMAIN IN COMPLEX WITH RAC-IV-016, a MK1775 analougue | | Descriptor: | 1,2-ETHANEDIOL, 6-{[4-(4-methylpiperazin-1-yl)phenyl]amino}-2-(prop-2-en-1-yl)-1-{6-[(1S)-2,2,2-trifluoro-1-hydroxyethyl]pyridin-2-yl}-1,2-dihydro-3H-pyrazolo[3,4-d]pyrimidin-3-one, CHLORIDE ION, ... | | Authors: | Zhu, J.-Y, Schonbrunn, E. | | Deposit date: | 2017-04-01 | | Release date: | 2018-04-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structural basis of Wee family kinase inhibition by small molecules
to be published
|
|
5VD8
 
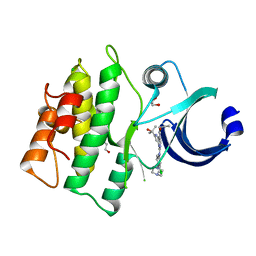 | | Crystal structure of human WEE1 kinase domain in complex with RAC-IV-099, a MK1775 analogue | | Descriptor: | 1,2-ETHANEDIOL, 6-{[3-chloro-4-(4-methylpiperazin-1-yl)phenyl]amino}-1-[6-(2-hydroxypropan-2-yl)pyridin-2-yl]-2-(prop-2-en-1-yl)-1,2-dihydro-3H-pyrazolo[3,4-d]pyrimidin-3-one, CHLORIDE ION, ... | | Authors: | Zhu, J.-Y, Schonbrunn, E. | | Deposit date: | 2017-04-01 | | Release date: | 2018-04-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis of Wee family kinase inhibition by small molecules
to be published
|
|
7T62
 
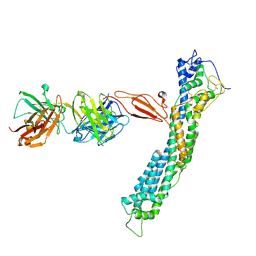 | | GPC2 HEP CT3 complex | | Descriptor: | CT3, Glypican-2 | | Authors: | Zhu, J, Cachau, R, De Val Alda, N, Li, N, Ho, M. | | Deposit date: | 2021-12-13 | | Release date: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (21 Å) | | Cite: | CAR T cells targeting tumor-associated exons of glypican 2 regress neuroblastoma in mice.
Cell Rep Med, 2, 2021
|
|
2IEL
 
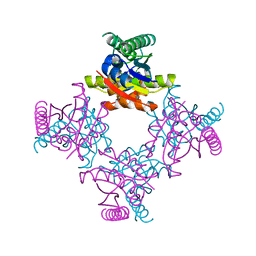 | | CRYSTAL STRUCTURE OF TT0030 from Thermus Thermophilus | | Descriptor: | Hypothetical Protein TT0030 | | Authors: | Zhu, J, Huang, J, Stepanyuk, G, Chen, L, Chang, J, Zhao, M, Xu, H, Liu, Z.J, Rose, J.P, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2006-09-19 | | Release date: | 2006-11-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | CRYSTAL STRUCTURE OF TT0030 from Thermus Thermophilus AT 1.6 ANGSTROMS RESOLUTION
To be Published
|
|
2KBK
 
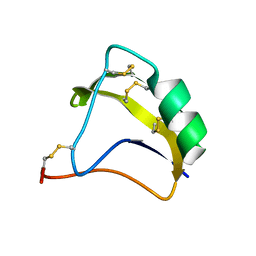 | | Solution Structure of BmK-M10 | | Descriptor: | Neurotoxin BmK-M10 | | Authors: | Zhu, J, Wu, H. | | Deposit date: | 2008-11-28 | | Release date: | 2009-12-22 | | Last modified: | 2019-12-11 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of BmK-M10
To be Published
|
|
2KBJ
 
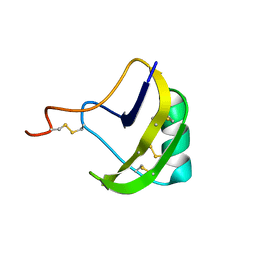 | | solution structure of BmKalphaTx11 (minor conformation) | | Descriptor: | Toxin Bmka2 | | Authors: | Zhu, J, Wu, H. | | Deposit date: | 2008-11-28 | | Release date: | 2009-12-08 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of BmKalphaTx11, a toxin from the venom of the Chinese scorpion Buthus martensii Karsch
Biochem.Biophys.Res.Commun., 391, 2010
|
|
2KBH
 
 | | solution structure of BmKalphaTx11 (major conformation) | | Descriptor: | Toxin Bmka2 | | Authors: | Zhu, J, Wu, H. | | Deposit date: | 2008-11-28 | | Release date: | 2009-12-08 | | Last modified: | 2019-12-11 | | Method: | SOLUTION NMR | | Cite: | Solution structure of BmKalphaTx11, a toxin from the venom of the Chinese scorpion Buthus martensii Karsch
Biochem.Biophys.Res.Commun., 391, 2010
|
|
