6M53
 
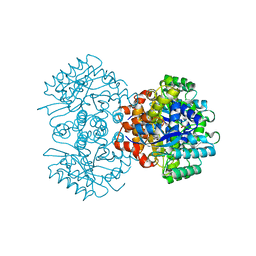 | | Crystal structure of 2, 3-dihydroxybenzoic acid decarboxylase from Fusarium oxysporum | | Descriptor: | 2,3-dihydroxybenzoate decarboxylase, GLYCEROL, ZINC ION | | Authors: | Song, M.K, Feng, J.H, Liu, W.D, Wu, Q.Q, Zhu, D.M. | | Deposit date: | 2020-03-09 | | Release date: | 2020-07-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | 2,3-Dihydroxybenzoic Acid Decarboxylase from Fusarium oxysporum: Crystal Structures and Substrate Recognition Mechanism.
Chembiochem, 21, 2020
|
|
8JQK
 
 | | Crystal structure of a carbonyl reductase SSCR mutant from Sporobolomyces Salmonicolor | | Descriptor: | Aldehyde reductase 2 | | Authors: | Zhang, H.L, Li, Q, Liu, W.D, Chen, X, Wu, Q.Q, Zhu, D.M. | | Deposit date: | 2023-06-14 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Engineering a Carbonyl Reductase to Simultaneously Increase Activity Toward Bulky Ketone and Isopropanol for Dynamic Kinetic Asymmetric Reduction via Enzymatic Hydrogen Transfer
Acs Catalysis, 13, 2023
|
|
8JQJ
 
 | | Crystal structure of carbonyl reductase SSCR mutant 1 from Sporobolomyces Salmonicolor | | Descriptor: | Aldehyde reductase 2 | | Authors: | Zhang, H.L, Li, Q, Liu, W.D, Chen, X, Wu, Q.Q, Zhu, D.M. | | Deposit date: | 2023-06-14 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Engineering a Carbonyl Reductase to Simultaneously Increase Activity Toward Bulky Ketone and Isopropanol for Dynamic Kinetic Asymmetric Reduction via Enzymatic Hydrogen Transfer
Acs Catalysis, 13, 2023
|
|
7BPC
 
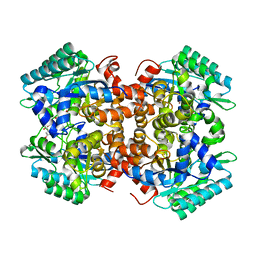 | | Crystal structure of 2, 3-dihydroxybenzoic acid decarboxylase from Fusarium oxysporum in complex with 2,5-DHBA | | Descriptor: | 2,3-dihydroxybenzoate decarboxylase, 2,5-dihydroxybenzoic acid, ZINC ION | | Authors: | Song, M.K, Feng, J.H, Liu, W.D, Wu, Q.Q, Zhu, D.M. | | Deposit date: | 2020-03-22 | | Release date: | 2020-07-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | 2,3-Dihydroxybenzoic Acid Decarboxylase from Fusarium oxysporum: Crystal Structures and Substrate Recognition Mechanism.
Chembiochem, 21, 2020
|
|
7BP1
 
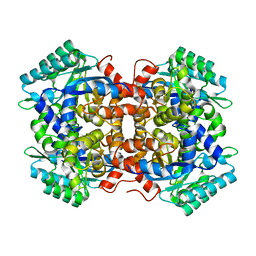 | | Crystal structure of 2, 3-dihydroxybenzoic acid decarboxylase from Fusarium oxysporum in complex with Catechol | | Descriptor: | 2,3-dihydroxybenzoate decarboxylase, CATECHOL, ZINC ION | | Authors: | Song, M.K, Feng, J.H, Liu, W.D, Wu, Q.Q, Zhu, D.M. | | Deposit date: | 2020-03-21 | | Release date: | 2020-07-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | 2,3-Dihydroxybenzoic Acid Decarboxylase from Fusarium oxysporum: Crystal Structures and Substrate Recognition Mechanism.
Chembiochem, 21, 2020
|
|
4WOQ
 
 | | Crystal Structures of CdNal from Clostridium difficile in complex with ketobutyric | | Descriptor: | 2-KETOBUTYRIC ACID, N-acetylneuraminate lyase | | Authors: | Liu, W.D, Guo, R.T, Cui, Y.F, Chen, X, Wu, Q.Q, Zhu, D.M. | | Deposit date: | 2014-10-16 | | Release date: | 2015-10-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structures of CdNal from Clostridium difficile in complex with ketobutyric
to be published
|
|
4WOZ
 
 | | Crystal Structures of CdNal from Clostridium difficile in complex with mannosamine | | Descriptor: | 2-(ACETYLAMINO)-2-DEOXY-D-MANNOSE, N-acetylneuraminate lyase | | Authors: | Liu, W.D, Guo, R.T, Cui, Y.F, Chen, X, Wu, Q.Q, Zhu, D.M. | | Deposit date: | 2014-10-17 | | Release date: | 2015-10-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal Structures of CdNal from Clostridium difficile in complex with mannosamine
to be published
|
|
8H8Y
 
 | | Crystal structure of AbHheG from Acidimicrobiia bacterium | | Descriptor: | GLYCEROL, alpha/beta hydrolase | | Authors: | Zhou, C.H, Chen, X, Han, X, Liu, W.D, Wu, Q.Q, Zhu, D.M, Ma, Y.H. | | Deposit date: | 2022-10-24 | | Release date: | 2023-08-02 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Flipping the Substrate Creates a Highly Selective Halohydrin Dehalogenase for the Synthesis of Chiral 4-Aryl-2-oxazolidinones from Readily Available Epoxides
Acs Catalysis, 13, 2023
|
|
8HQP
 
 | | Crystal structure of AbHheG mutant from Acidimicrobiia bacterium | | Descriptor: | AbHheG_m | | Authors: | Zhou, C.H, Chen, X, Han, X, Liu, W.D, Wu, Q.Q, Zhu, D.M, Ma, Y.H. | | Deposit date: | 2022-12-13 | | Release date: | 2023-08-02 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Flipping the Substrate Creates a Highly Selective Halohydrin Dehalogenase for the Synthesis of Chiral 4-Aryl-2-oxazolidinones from Readily Available Epoxides
Acs Catalysis, 13, 2023
|
|
6IHH
 
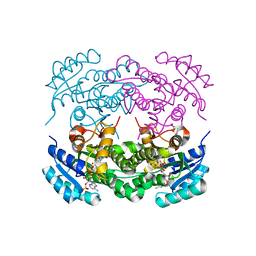 | | Crystal structure of RasADH F12 from Ralstonia.sp in complex with NADPH and A6O | | Descriptor: | (2R,3S)-2-ethyl-2-[(2E)-2-(6-methoxy-3,4-dihydro-2H-naphthalen-1-ylidene)ethyl]-3-oxidanyl-cyclopentan-1-one, Alclohol dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zhang, H.L, Chen, X, Liu, W.D, Wu, Q.Q, Zhu, D.M. | | Deposit date: | 2018-09-30 | | Release date: | 2019-10-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Efficient reductive desymmetrization of bulky 1,3-cyclodiketones enabled by structure-guided directed evolution of a carbonyl reductase.
Nat Catal, 2, 2019
|
|
6IHI
 
 | | Crystal structure of RasADH 3B3/I91V from Ralstonia.sp in complex with NADPH and A6O | | Descriptor: | (2R,3S)-2-ethyl-2-[(2E)-2-(6-methoxy-3,4-dihydro-2H-naphthalen-1-ylidene)ethyl]-3-oxidanyl-cyclopentan-1-one, Alclohol dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zhang, H.L, Chen, X, Liu, W.D, Wu, Q.Q, Zhu, D.M. | | Deposit date: | 2018-09-30 | | Release date: | 2019-10-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Efficient reductive desymmetrization of bulky 1,3-cyclodiketones enabled by structure-guided directed evolution of a carbonyl reductase.
Nat Catal, 2, 2019
|
|
3WGQ
 
 | | Crystal structure of meso-dapdh Q154L/T173I/R199M/P248S/H249N/N276S mutant with DAP of from Clostridium tetani E88 | | Descriptor: | 2,6-DIAMINOPIMELIC ACID, Meso-diaminopimelate dehydrogenase | | Authors: | Liu, W.D, Li, Z, Huang, C.H, Guo, R.T, Wu, Q.Q, Zhu, D.M. | | Deposit date: | 2013-08-07 | | Release date: | 2014-08-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of meso-dapdh Q154L/T173I/R199M/P248S/H249N/N276S mutant with DAP of from Clostridium tetani E88
To be Published
|
|
3WGZ
 
 | | Crystal structure of meso-dapdh Q154L/T173I/R199M/P248S/H249N/N276S mutant with D-leucine of from Clostridium tetani E88 | | Descriptor: | D-LEUCINE, Meso-diaminopimelate dehydrogenase | | Authors: | Liu, W.D, Li, Z, Huang, C.H, Guo, R.T, Wu, Q.Q, Zhu, D.M. | | Deposit date: | 2013-08-20 | | Release date: | 2014-08-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of meso-dapdh Q154L/T173I/R199M/P248S/H249N/N276S mutant with D-leucine of from Clostridium tetani E88
to be published
|
|
3WGO
 
 | | Crystal structure of Q154L/T173I/R199M/P248S/H249/N276S mutant of meso-dapdh from Clostridium tetani E88 | | Descriptor: | Meso-diaminopimelate dehydrogenase | | Authors: | Liu, W.D, Li, Z, Huang, C.H, Guo, R.T, Wu, Q.Q, Zhu, D.M. | | Deposit date: | 2013-08-06 | | Release date: | 2014-08-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of Q154L/T173I/R199M/P248S/H249 mutant of meso-dapdh from Clostridium tetani E88
To be Published
|
|
3WGY
 
 | | Crystal structure of meso-dapdh Q154L/T173I/R199M/P248S/H249N/N276S mutant with 4-methyl-2-oxovalerate of from Clostridium tetani E88 | | Descriptor: | 2-OXO-4-METHYLPENTANOIC ACID, Meso-diaminopimelate dehydrogenase | | Authors: | Liu, W.D, Li, Z, Huang, C.H, Guo, R.T, Wu, Q.Q, Zhu, D.M. | | Deposit date: | 2013-08-19 | | Release date: | 2014-08-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of meso-dapdh Q154L/T173I/R199M/P248S/H249N/N276S mutant with 4-methyl-2-oxovalerate of from Clostridium tetani E88
to be published
|
|
7DL0
 
 | | The mutant E310G/A314Y of 3,5-DAHDHcca complex with NADPH | | Descriptor: | 1,2-ETHANEDIOL, 3,5-diaminohexanoate dehydrogenase, CHLORIDE ION, ... | | Authors: | Liu, N, Wu, L, Zhu, D.M, Zhou, J.H. | | Deposit date: | 2020-11-25 | | Release date: | 2021-09-29 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Crystal Structures and Catalytic Mechanism of l-erythro-3,5-Diaminohexanoate Dehydrogenase and Rational Engineering for Asymmetric Synthesis of beta-Amino Acids.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7DL7
 
 | | The wild-type structure of 3,5-DAHDHcca | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3,5-diaminohexanoate dehydrogenase, ... | | Authors: | Liu, N, Wu, L, Zhu, D.M, Zhou, J.H. | | Deposit date: | 2020-11-26 | | Release date: | 2021-09-29 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.30065823 Å) | | Cite: | Crystal Structures and Catalytic Mechanism of l-erythro-3,5-Diaminohexanoate Dehydrogenase and Rational Engineering for Asymmetric Synthesis of beta-Amino Acids.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7DL1
 
 | | The mutant E310G/G323S structure of 3,5-DAHDHcca complex with NADPH | | Descriptor: | 3,5-diaminohexanoate dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Liu, N, Wu, L, Zhu, D.M, Zhou, J.H. | | Deposit date: | 2020-11-25 | | Release date: | 2021-09-29 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Crystal Structures and Catalytic Mechanism of l-erythro-3,5-Diaminohexanoate Dehydrogenase and Rational Engineering for Asymmetric Synthesis of beta-Amino Acids.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7DL3
 
 | | The structure of 3,5-DAHDHcca complex with NADPH | | Descriptor: | 3,5-diaminohexanoate dehydrogenase, CHLORIDE ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Liu, N, Wu, L, Zhu, D.M, Zhou, J.H. | | Deposit date: | 2020-11-25 | | Release date: | 2021-09-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.84606934 Å) | | Cite: | Crystal Structures and Catalytic Mechanism of l-erythro-3,5-Diaminohexanoate Dehydrogenase and Rational Engineering for Asymmetric Synthesis of beta-Amino Acids.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
3WBF
 
 | | Crystal Structure of meso-diaminopimelate dehydrogenase from Symbiobacterium thermophilum co-crystallized with NADP+ and DAP | | Descriptor: | 2,6-DIAMINOPIMELIC ACID, Diaminopimelate dehydrogenase, GLYCEROL, ... | | Authors: | Liu, W.D, Li, Z, Huang, C.H, Guo, R.T, Wu, Q.Q, Zhu, D.M. | | Deposit date: | 2013-05-15 | | Release date: | 2014-03-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structural and mutational studies on the unusual substrate specificity of meso-diaminopimelate dehydrogenase from Symbiobacterium thermophilum.
Chembiochem, 15, 2014
|
|
3WB9
 
 | | Crystal Structures of meso-diaminopimelate dehydrogenase from Symbiobacterium thermophilum | | Descriptor: | Diaminopimelate dehydrogenase, GLYCEROL | | Authors: | Liu, W.D, Li, Z, Huang, C.H, Guo, R.T, Wu, Q.Q, Zhu, D.M. | | Deposit date: | 2013-05-14 | | Release date: | 2014-03-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural and mutational studies on the unusual substrate specificity of meso-diaminopimelate dehydrogenase from Symbiobacterium thermophilum.
Chembiochem, 15, 2014
|
|
3WBB
 
 | | Crystal Structures of meso-diaminopimelate dehydrogenase from Symbiobacterium thermophilum | | Descriptor: | Diaminopimelate dehydrogenase, GLYCEROL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Liu, W.D, Li, Z, Huang, C.H, Guo, R.T, Wu, Q.Q, Zhu, D.M. | | Deposit date: | 2013-05-14 | | Release date: | 2014-03-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural and mutational studies on the unusual substrate specificity of meso-diaminopimelate dehydrogenase from Symbiobacterium thermophilum.
Chembiochem, 15, 2014
|
|
