7WXH
 
 | | GPR domain open form of Drosophila P5CS filament with glutamate, ATP, and NADPH | | Descriptor: | Delta-1-pyrroline-5-carboxylate synthase | | Authors: | Liu, J.L, Zhong, J, Guo, C.J, Zhou, X. | | Deposit date: | 2022-02-14 | | Release date: | 2022-03-30 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural basis of dynamic P5CS filaments.
Elife, 11, 2022
|
|
7WXG
 
 | | GPR domain closed form of Drosophila P5CS filament with glutamate, ATP, and NADPH | | Descriptor: | Delta-1-pyrroline-5-carboxylate synthase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Liu, J.L, Zhong, J, Guo, C.J, Zhou, X. | | Deposit date: | 2022-02-14 | | Release date: | 2022-03-30 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural basis of dynamic P5CS filaments.
Elife, 11, 2022
|
|
7WX4
 
 | |
7WX3
 
 | | GK domain of Drosophila P5CS filament with glutamate, ATP, and NADPH | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Delta-1-pyrroline-5-carboxylate synthase, GAMMA-GLUTAMYL PHOSPHATE, ... | | Authors: | Liu, J.L, Zhong, J, Guo, C.J, Zhou, X. | | Deposit date: | 2022-02-14 | | Release date: | 2022-04-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of dynamic P5CS filaments.
Elife, 11, 2022
|
|
7WD4
 
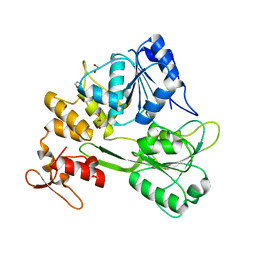 | | Crystal structure of the Ilheus virus helicase: implications for enzyme function and drug design | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, NS3 helicase | | Authors: | Wang, D.P, Wang, M.Y, Zhou, X, Wang, W.M, Cao, J.M. | | Deposit date: | 2021-12-21 | | Release date: | 2023-01-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of the Ilheus virus helicase: implications for enzyme function and drug design
To Be Published
|
|
8HUZ
 
 | |
7CYG
 
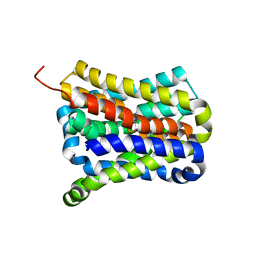 | | Crystal structure of a cysteine-pair mutant (Y113C-P190C) of a bacterial bile acid transporter before disulfide bond formation | | Descriptor: | Transporter, sodium/bile acid symporter family | | Authors: | Wang, X, Lyu, Y, Ji, Y, Sun, Z, Zhou, X. | | Deposit date: | 2020-09-03 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.198 Å) | | Cite: | An engineered disulfide bridge traps and validates an outward-facing conformation in a bile acid transporter.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7CYK
 
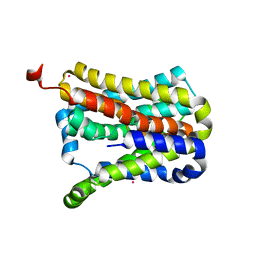 | | Crystal structure of a second cysteine-pair mutant (V110C-I197C) of a bacterial bile acid transporter before disulfide bond formation | | Descriptor: | MERCURY (II) ION, Transporter, sodium/bile acid symporter family | | Authors: | Wang, X, Lyu, Y, Ji, Y, Sun, Z, Zhou, X. | | Deposit date: | 2020-09-03 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.785 Å) | | Cite: | An engineered disulfide bridge traps and validates an outward-facing conformation in a bile acid transporter.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7DIX
 
 | | Crystal structure of LeuT in lipidic cubic phase at pH 5 | | Descriptor: | Na(+):neurotransmitter symporter (Snf family), SELENOMETHIONINE, SODIUM ION | | Authors: | Fan, J, Xiao, Y, Sun, Z, Zhou, X. | | Deposit date: | 2020-11-19 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.49 Å) | | Cite: | Crystal structures of LeuT reveal conformational dynamics in the outward-facing states.
J.Biol.Chem., 296, 2021
|
|
7DJC
 
 | | Crystal structure of the G26C/Q250A mutant of LeuT | | Descriptor: | LEUCINE, Na(+):neurotransmitter symporter (Snf family), SODIUM ION, ... | | Authors: | Fan, J, Xiao, Y, Sun, Z, Zhou, X. | | Deposit date: | 2020-11-20 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | Crystal structures of LeuT reveal conformational dynamics in the outward-facing states.
J.Biol.Chem., 296, 2021
|
|
7DJ2
 
 | | Crystal structure of the G26C/E290S mutant of LeuT | | Descriptor: | LEUCINE, Na(+):neurotransmitter symporter (Snf family), SODIUM ION, ... | | Authors: | Fan, J, Xiao, Y, Sun, Z, Zhou, X. | | Deposit date: | 2020-11-19 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of LeuT reveal conformational dynamics in the outward-facing states.
J.Biol.Chem., 296, 2021
|
|
7DJ1
 
 | | Crystal structure of the G26C mutant of LeuT | | Descriptor: | LEUCINE, Na(+):neurotransmitter symporter (Snf family), SODIUM ION | | Authors: | Fan, J, Xiao, Y, Sun, Z, Zhou, X. | | Deposit date: | 2020-11-19 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.528 Å) | | Cite: | Crystal structures of LeuT reveal conformational dynamics in the outward-facing states.
J.Biol.Chem., 296, 2021
|
|
7DII
 
 | | Crystal structure of LeuT in lipidic cubic phase at pH 7 | | Descriptor: | LEUCINE, Na(+):neurotransmitter symporter (Snf family), SODIUM ION | | Authors: | Fan, J, Xiao, Y, Sun, Z, Zhou, X. | | Deposit date: | 2020-11-19 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.403 Å) | | Cite: | Crystal structures of LeuT reveal conformational dynamics in the outward-facing states.
J.Biol.Chem., 296, 2021
|
|
7F5X
 
 | | GK domain of Drosophila P5CS filament with glutamate | | Descriptor: | Delta-1-pyrroline-5-carboxylate synthase, GAMMA-L-GLUTAMIC ACID | | Authors: | Liu, J.L, Zhong, J, Guo, C.J, Zhou, X. | | Deposit date: | 2021-06-23 | | Release date: | 2022-04-06 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis of dynamic P5CS filaments.
Elife, 11, 2022
|
|
7F5V
 
 | | Drosophila P5CS filament with glutamate, ATP, and NADPH | | Descriptor: | Delta-1-pyrroline-5-carboxylate synthase | | Authors: | Liu, J.L, Zhong, J, Guo, C.J, Zhou, X. | | Deposit date: | 2021-06-22 | | Release date: | 2022-04-06 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis of dynamic P5CS filaments.
Elife, 11, 2022
|
|
7F5U
 
 | |
7F5T
 
 | | Drosophila P5CS filament with glutamate | | Descriptor: | Delta-1-pyrroline-5-carboxylate synthase, GLUTAMIC ACID | | Authors: | Liu, J.L, Zhong, J, Guo, C.J, Zhou, X. | | Deposit date: | 2021-06-22 | | Release date: | 2022-05-18 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural basis of dynamic P5CS filaments.
Elife, 11, 2022
|
|
7C72
 
 | | Structure of a mycobacterium tuberculosis puromycin-hydrolyzing peptidase | | Descriptor: | D-MALATE, GLYCEROL, Prolyl oligopeptidase | | Authors: | Ruiz-Carrillo, D, Zhao, Y.H, Feng, Q, Zhou, X, Zhang, Y, Jiang, J, Lukman, M. | | Deposit date: | 2020-05-22 | | Release date: | 2021-03-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.00004458 Å) | | Cite: | Mycobacterium tuberculosis puromycin hydrolase displays a prolyl oligopeptidase fold and an acyl aminopeptidase activity.
Proteins, 89, 2021
|
|
7DPT
 
 | | Structural basis for ligand binding modes of CTP synthase | | Descriptor: | 6-DIAZENYL-5-OXO-L-NORLEUCINE, ADENOSINE-5'-DIPHOSPHATE, CTP synthase, ... | | Authors: | Liu, J.L, Zhou, X, Guo, C.J, Chang, C.C. | | Deposit date: | 2020-12-21 | | Release date: | 2021-09-15 | | Method: | ELECTRON MICROSCOPY (2.48 Å) | | Cite: | Structural basis for ligand binding modes of CTP synthase.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7DPW
 
 | | Structural basis for ligand binding modes of CTP synthase | | Descriptor: | CTP synthase, CYTIDINE-5'-TRIPHOSPHATE, MAGNESIUM ION | | Authors: | Liu, J.L, Zhou, X, Guo, C.J, Chang, C.C. | | Deposit date: | 2020-12-21 | | Release date: | 2021-09-15 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.65 Å) | | Cite: | Structural basis for ligand binding modes of CTP synthase.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
