8DLP
 
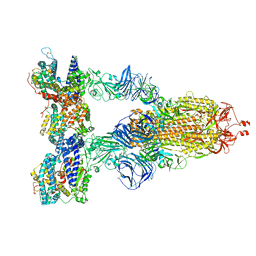 | | Cryo-EM structure of SARS-CoV-2 Gamma (P.1) spike protein in complex with human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Zhu, X, Mannar, D, Saville, J.W, Srivastava, S.S, Berezuk, A.M, Zhou, S, Tuttle, K.S, Subramaniam, S. | | Deposit date: | 2022-07-08 | | Release date: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (2.64 Å) | | Cite: | SARS-CoV-2 variants of concern: spike protein mutational analysis and epitope for broad neutralization.
Nat Commun, 13, 2022
|
|
8DLX
 
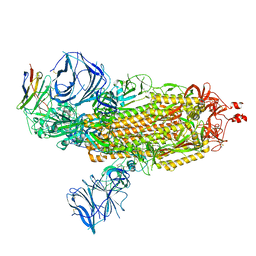 | | Cryo-EM structure of SARS-CoV-2 Epsilon (B.1.429) spike protein in complex with VH ab6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Zhu, X, Mannar, D, Saville, J.W, Srivastava, S.S, Berezuk, A.M, Zhou, S, Tuttle, K.S, Subramaniam, S. | | Deposit date: | 2022-07-08 | | Release date: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (2.45 Å) | | Cite: | SARS-CoV-2 variants of concern: spike protein mutational analysis and epitope for broad neutralization.
Nat Commun, 13, 2022
|
|
8DM0
 
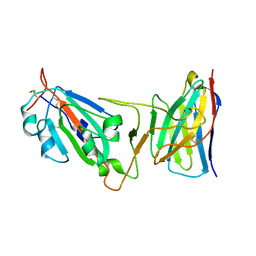 | | Cryo-EM structure of SARS-CoV-2 D614G spike protein in complex with VH ab6 (focused refinement of NTD and VH ab6) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, VH ab6 | | Authors: | Zhu, X, Mannar, D, Saville, J.W, Srivastava, S.S, Berezuk, A.M, Zhou, S, Tuttle, K.S, Subramaniam, S. | | Deposit date: | 2022-07-08 | | Release date: | 2022-08-31 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | SARS-CoV-2 variants of concern: spike protein mutational analysis and epitope for broad neutralization.
Nat Commun, 13, 2022
|
|
8DLM
 
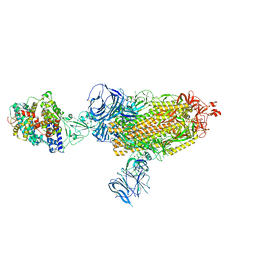 | | Cryo-EM structure of SARS-CoV-2 Beta (B.1.351) spike protein in complex with human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Zhu, X, Mannar, D, Saville, J.W, Srivastava, S.S, Berezuk, A.M, Zhou, S, Tuttle, K.S, Subramaniam, S. | | Deposit date: | 2022-07-08 | | Release date: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | SARS-CoV-2 variants of concern: spike protein mutational analysis and epitope for broad neutralization.
Nat Commun, 13, 2022
|
|
8DLL
 
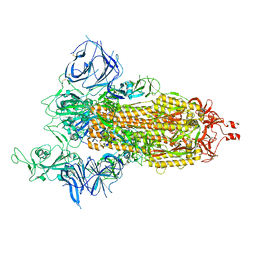 | | Cryo-EM structure of SARS-CoV-2 Beta (B.1.351) spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhu, X, Mannar, D, Saville, J.W, Srivastava, S.S, Berezuk, A.M, Zhou, S, Tuttle, K.S, Subramaniam, S. | | Deposit date: | 2022-07-08 | | Release date: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (2.56 Å) | | Cite: | SARS-CoV-2 variants of concern: spike protein mutational analysis and epitope for broad neutralization.
Nat Commun, 13, 2022
|
|
8DLI
 
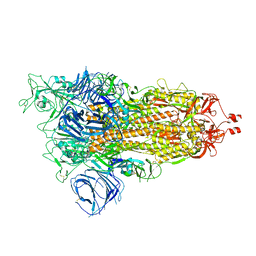 | | Cryo-EM structure of SARS-CoV-2 Alpha (B.1.1.7) spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhu, X, Mannar, D, Saville, J.W, Srivastava, S.S, Berezuk, A.M, Zhou, S, Tuttle, K.S, Subramaniam, S. | | Deposit date: | 2022-07-08 | | Release date: | 2022-08-31 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.56 Å) | | Cite: | SARS-CoV-2 variants of concern: spike protein mutational analysis and epitope for broad neutralization.
Nat Commun, 13, 2022
|
|
8DLQ
 
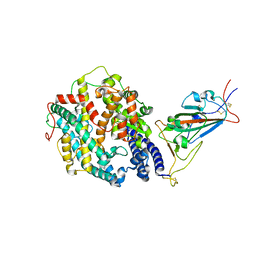 | | Cryo-EM structure of SARS-CoV-2 Gamma (P.1) spike protein in complex with human ACE2 (focused refinement of RBD and ACE2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Zhu, X, Mannar, D, Saville, J.W, Srivastava, S.S, Berezuk, A.M, Zhou, S, Tuttle, K.S, Subramaniam, S. | | Deposit date: | 2022-07-08 | | Release date: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | SARS-CoV-2 variants of concern: spike protein mutational analysis and epitope for broad neutralization.
Nat Commun, 13, 2022
|
|
8DLZ
 
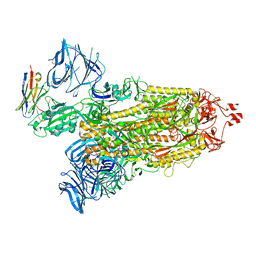 | | Cryo-EM structure of SARS-CoV-2 D614G spike protein in complex with VH ab6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Zhu, X, Mannar, D, Saville, J.W, Srivastava, S.S, Berezuk, A.M, Zhou, S, Tuttle, K.S, Subramaniam, S. | | Deposit date: | 2022-07-08 | | Release date: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (2.57 Å) | | Cite: | SARS-CoV-2 variants of concern: spike protein mutational analysis and epitope for broad neutralization.
Nat Commun, 13, 2022
|
|
3OWG
 
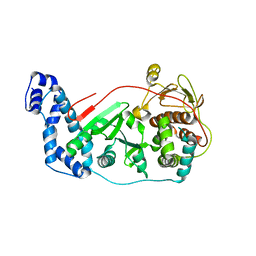 | | Crystal structure of vaccinia virus Polyadenylate polymerase(vp55) | | Descriptor: | Poly(A) polymerase catalytic subunit | | Authors: | Li, C, Li, H, Zhou, S, Gershon, P.D, Poulos, T.L. | | Deposit date: | 2010-09-17 | | Release date: | 2011-09-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Domain-level rocking motion within a polymerase that translocates on single-stranded nucleic acid.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3ERC
 
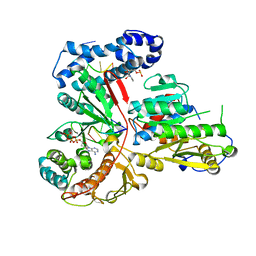 | | Crystal structure of the heterodimeric vaccinia virus mRNA polyadenylate polymerase with three fragments of RNA and 3'-deoxy ATP | | Descriptor: | 3'-DEOXYADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, Cap-specific mRNA (nucleoside-2'-O-)-methyltransferase, ... | | Authors: | Li, C, Li, H, Zhou, S, Poulos, T.L, Gershon, P.D. | | Deposit date: | 2008-10-01 | | Release date: | 2009-06-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | Polymerase Translocation with Respect to Single-Stranded Nucleic Acid: Looping or Wrapping of Primer around a Poly(A) Polymerase
Structure, 17, 2009
|
|
3ER9
 
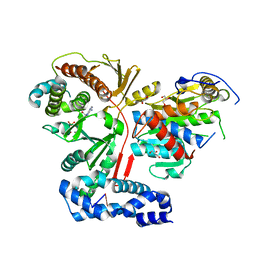 | | Crystal structure of the heterodimeric vaccinia virus mRNA polyadenylate polymerase complex with UU and 3'-deoxy ATP | | Descriptor: | 3'-DEOXYADENOSINE-5'-TRIPHOSPHATE, 5'-R(UP*U)-3', CALCIUM ION, ... | | Authors: | Li, C, Li, H, Zhou, S, Poulos, T.L, Gershon, P.D. | | Deposit date: | 2008-10-01 | | Release date: | 2009-06-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Polymerase Translocation with Respect to Single-Stranded Nucleic Acid: Looping or Wrapping of Primer around a Poly(A) Polymerase
Structure, 17, 2009
|
|
3ER8
 
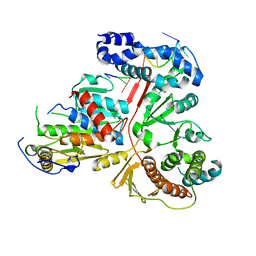 | | Crystal structure of the heterodimeric vaccinia virus mRNA polyadenylate polymerase complex with two fragments of RNA | | Descriptor: | Cap-specific mRNA (nucleoside-2'-O-)-methyltransferase, Poly(A) polymerase catalytic subunit, RNA/DNA chimera 5'-D(CP*CP*)R(UP*UP*)D(C)-3', ... | | Authors: | Li, C, Li, H, Zhou, S, Poulos, T.L, Gershon, P.D. | | Deposit date: | 2008-10-01 | | Release date: | 2009-06-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | Polymerase Translocation with Respect to Single-Stranded Nucleic Acid: Looping or Wrapping of Primer around a Poly(A) Polymerase
Structure, 17, 2009
|
|
5NO4
 
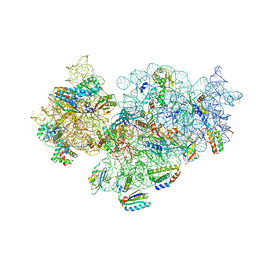 | | RsgA-GDPNP bound to the 30S ribosomal subunit (RsgA assembly intermediate with uS3) | | Descriptor: | 16S ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Lopez-Alonso, J.P, Kaminishi, T, Kikuchi, T, Hirata, Y, Iturrioz, I, Dhimole, N, Schedlbauer, A, Hase, Y, Goto, S, Kurita, D, Muto, A, Zhou, S, Naoe, C, Mills, D.J, Gil-Carton, D, Takemoto, C, Himeno, H, Fucini, P, Connell, S.R. | | Deposit date: | 2017-04-10 | | Release date: | 2017-05-31 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (5.16 Å) | | Cite: | RsgA couples the maturation state of the 30S ribosomal decoding center to activation of its GTPase pocket.
Nucleic Acids Res., 45, 2017
|
|
5NO2
 
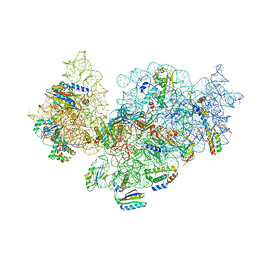 | | RsgA-GDPNP bound to the 30S ribosomal subunit (RsgA assembly intermediate) | | Descriptor: | 16S ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Lopez-Alonso, J.P, Kaminishi, T, Kikuchi, T, Hirata, Y, Iturrioz, I, Dhimole, N, Schedlbauer, A, Hase, Y, Goto, S, Kurita, D, Muto, A, Zhou, S, Naoe, C, Mills, D.J, Gil-Carton, D, Takemoto, C, Himeno, H, Fucini, P, Connell, S.R. | | Deposit date: | 2017-04-10 | | Release date: | 2017-05-24 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (5.16 Å) | | Cite: | RsgA couples the maturation state of the 30S ribosomal decoding center to activation of its GTPase pocket.
Nucleic Acids Res., 45, 2017
|
|
5NO3
 
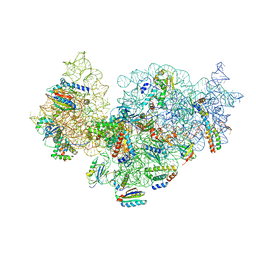 | | RsgA-GDPNP bound to the 30S ribosomal subunit (RsgA assembly intermediate without uS3) | | Descriptor: | 16S ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Lopez-Alonso, J.P, Kaminishi, T, Kikuchi, T, Hirata, Y, Iturrioz, I, Dhimole, N, Schedlbauer, A, Hase, Y, Goto, S, Kurita, D, Muto, A, Zhou, S, Naoe, C, Mills, D.J, Gil-Carton, D, Takemoto, C, Himeno, H, Fucini, P, Connell, S.R. | | Deposit date: | 2017-04-10 | | Release date: | 2017-05-31 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (5.16 Å) | | Cite: | RsgA couples the maturation state of the 30S ribosomal decoding center to activation of its GTPase pocket.
Nucleic Acids Res., 45, 2017
|
|
1XZY
 
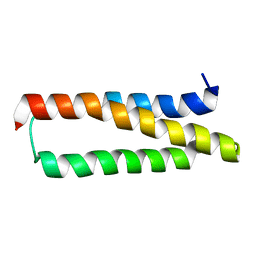 | | Solution structure of the P30-trans form of Alpha Hemoglobin Stabilizing Protein (AHSP) | | Descriptor: | Alpha-hemoglobin stabilizing protein | | Authors: | Gell, D.A, Feng, L, Zhou, S, Kong, Y, Lee, C, Weiss, M.J, Shi, Y, Mackay, J.P. | | Deposit date: | 2004-11-12 | | Release date: | 2004-12-21 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Molecular mechanism of AHSP-mediated stabilization of alpha-hemoglobin
Cell(Cambridge,Mass.), 119, 2004
|
|
1Y01
 
 | | Crystal structure of AHSP bound to Fe(II) alpha-hemoglobin | | Descriptor: | 6-[(CYCLOHEXYLACETYL)(2-HYDROXYETHYL)AMINO]-6-DEOXY-D-XYLO-HEXITOL, Alpha-hemoglobin stabilizing protein, Hemoglobin alpha chain, ... | | Authors: | Feng, L, Gell, D.A, Zhou, S, Gu, L, Gow, A.J, Weiss, M.J, Mackay, J.P, Shi, Y. | | Deposit date: | 2004-11-14 | | Release date: | 2004-12-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular mechanism of AHSP-mediated stabilization of alpha-hemoglobin.
Cell(Cambridge,Mass.), 119, 2004
|
|
1Z8U
 
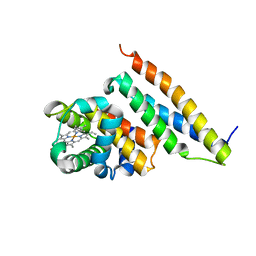 | | Crystal structure of oxidized alpha hemoglobin bound to AHSP | | Descriptor: | Alpha-hemoglobin stabilizing protein, Hemoglobin alpha chain, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Feng, L, Zhou, S, Gu, L, Gell, D.A, Mackay, J.P, Weiss, M.J, Gow, A.J, Shi, Y. | | Deposit date: | 2005-03-31 | | Release date: | 2005-06-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of oxidized alpha-haemoglobin bound to AHSP reveals a protective mechanism for haem.
Nature, 435, 2005
|
|
4YHH
 
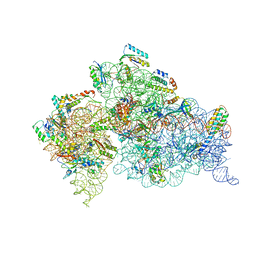 | | Crystal structure of the 30S ribosomal subunit from Thermus thermophilus in complex with tigecycline | | Descriptor: | 16S ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Schedlbauer, A, Kaminishi, T, Ochoa-Lizarralde, B, Dhimole, N, Zhou, S, Lopez-Alonso, J.P, Connell, S.R, Fucini, P. | | Deposit date: | 2015-02-27 | | Release date: | 2015-04-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.417 Å) | | Cite: | Structural characterization of an alternative mode of tigecycline binding to the bacterial ribosome.
Antimicrob.Agents Chemother., 59, 2015
|
|
1FOA
 
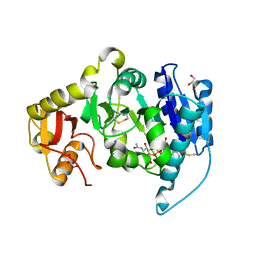 | | CRYSTAL STRUCTURE OF N-ACETYLGLUCOSAMINYLTRANSFERASE I | | Descriptor: | ALPHA-1,3-MANNOSYL-GLYCOPROTEIN BETA-1,2-N-ACETYLGLUCOSAMINYLTRANSFERASE, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Unligil, U.M, Zhou, S, Yuwaraj, S, Sarkar, M, Schachter, H, Rini, J.M. | | Deposit date: | 2000-08-26 | | Release date: | 2001-05-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray crystal structure of rabbit N-acetylglucosaminyltransferase I: catalytic mechanism and a new protein superfamily.
EMBO J., 19, 2000
|
|
1FO8
 
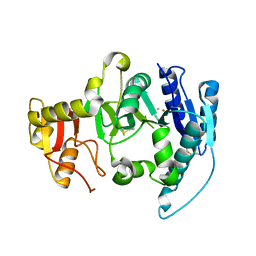 | | CRYSTAL STRUCTURE OF N-ACETYLGLUCOSAMINYLTRANSFERASE I | | Descriptor: | ALPHA-1,3-MANNOSYL-GLYCOPROTEIN BETA-1,2-N-ACETYLGLUCOSAMINYLTRANSFERASE, METHYL MERCURY ION | | Authors: | Unligil, U.M, Zhou, S, Yuwaraj, S, Sarkar, M, Schachter, H, Rini, J.M. | | Deposit date: | 2000-08-26 | | Release date: | 2001-04-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | X-ray crystal structure of rabbit N-acetylglucosaminyltransferase I: catalytic mechanism and a new protein superfamily.
EMBO J., 19, 2000
|
|
1FO9
 
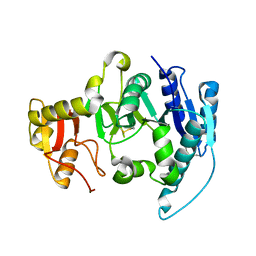 | | CRYSTAL STRUCTURE OF N-ACETYLGLUCOSAMINYLTRANSFERASE I | | Descriptor: | ALPHA-1,3-MANNOSYL-GLYCOPROTEIN BETA-1,2-N-ACETYLGLUCOSAMINYLTRANSFERASE | | Authors: | Unligil, U.M, Zhou, S, Yuwaraj, S, Sarkar, M, Schachter, H, Rini, J.M. | | Deposit date: | 2000-08-26 | | Release date: | 2001-04-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | X-ray crystal structure of rabbit N-acetylglucosaminyltransferase I: catalytic mechanism and a new protein superfamily.
EMBO J., 19, 2000
|
|
1J4Q
 
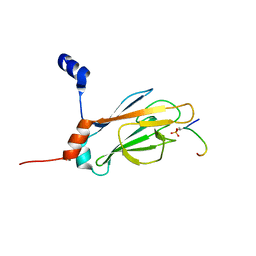 | | NMR STRUCTURE OF THE FHA1 DOMAIN OF RAD53 IN COMPLEX WITH A RAD9-DERIVED PHOSPHOTHREONINE (AT T192) PEPTIDE | | Descriptor: | DNA REPAIR PROTEIN RAD9, PROTEIN KINASE SPK1 | | Authors: | Yuan, C, Yongkiettrakul, S, Byeon, I.-J.L, Zhou, S, Tsai, M.-D. | | Deposit date: | 2001-10-22 | | Release date: | 2001-12-05 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Solution structures of two FHA1-phosphothreonine peptide complexes provide insight into the structural basis of the ligand specificity of FHA1 from yeast Rad53.
J.Mol.Biol., 314, 2001
|
|
1J4P
 
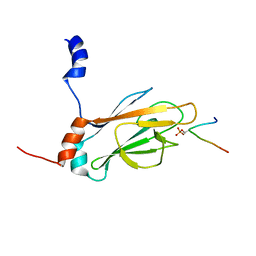 | | NMR STRUCTURE OF THE FHA1 DOMAIN OF RAD53 IN COMPLEX WITH A RAD9-DERIVED PHOSPHOTHREONINE (AT T155) PEPTIDE | | Descriptor: | DNA REPAIR PROTEIN RAD9, PROTEIN KINASE SPK1 | | Authors: | Yuan, C, Yongkiettrakul, S, Byeon, I.-J.L, Zhou, S, Tsai, M.-D. | | Deposit date: | 2001-10-22 | | Release date: | 2001-12-05 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Solution structures of two FHA1-phosphothreonine peptide complexes provide insight into the structural basis of the ligand specificity of FHA1 from yeast Rad53.
J.Mol.Biol., 314, 2001
|
|
1K3N
 
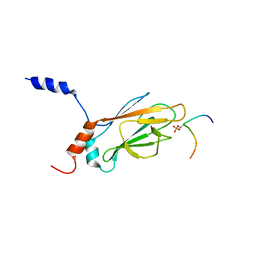 | | NMR Structure of the FHA1 Domain of Rad53 in Complex with a Rad9-derived Phosphothreonine (at T155) Peptide | | Descriptor: | DNA repair protein Rad9, Protein Kinase SPK1 | | Authors: | Yuan, C, Yongkiettrakul, S, Byeon, I.-J.L, Zhou, S, Tsai, M.-D. | | Deposit date: | 2001-10-03 | | Release date: | 2001-12-05 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structures of two FHA1-phosphothreonine peptide complexes provide insight into the structural basis of the ligand specificity of FHA1 from yeast Rad53.
J.Mol.Biol., 314, 2001
|
|
