1HU9
 
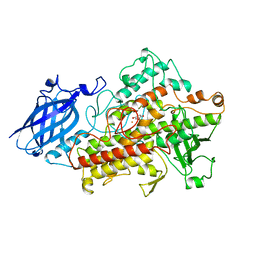 | | LIPOXYGENASE-3 (SOYBEAN) COMPLEX WITH 4-HYDROPEROXY-2-METHOXY-PHENOL | | Descriptor: | 4-HYDROPEROXY-2-METHOXY-PHENOL, FE (III) ION, LIPOXYGENASE-3 | | Authors: | Zhou, K, Skrzypczak-Jankun, E, McCabe, N.P, Selman, S.H, Jankun, J. | | Deposit date: | 2001-01-04 | | Release date: | 2003-06-03 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of curcumin in complex with lipoxygenase and its significance in
cancer.
INT.J.MOL.MED., 12, 2003
|
|
6C0W
 
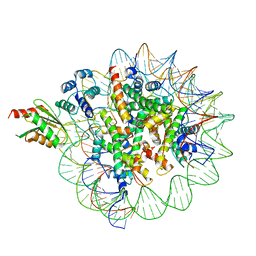 | | Cryo-EM structure of human kinetochore protein CENP-N with the centromeric nucleosome containing CENP-A | | Descriptor: | 147 mer DNA, Centromere protein N, Histone H2A, ... | | Authors: | Zhou, K, Pentakota, S, Vetter, I.R, Morgan, G.P, Petrovic, A, Musacchio, A, Luger, K. | | Deposit date: | 2018-01-02 | | Release date: | 2018-01-17 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Decoding the centromeric nucleosome through CENP-N.
Elife, 6, 2017
|
|
6VRA
 
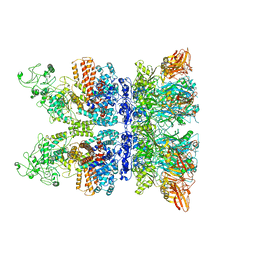 | | Anthrax octamer prechannel bound to full-length edema factor | | Descriptor: | CALCIUM ION, Calmodulin-sensitive adenylate cyclase, Protective antigen | | Authors: | Zhou, K, Hardenbrook, N.J, Liu, S, Cui, Y.X, Krantz, B.A, Zhou, Z.H. | | Deposit date: | 2020-02-07 | | Release date: | 2020-12-16 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Atomic Structures of Anthrax Prechannel Bound with Full-Length Lethal and Edema Factors.
Structure, 28, 2020
|
|
6WJJ
 
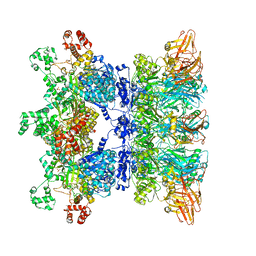 | | Anthrax octamer prechannel bound to full-length lethal factor | | Descriptor: | CALCIUM ION, Lethal factor, Protective antigen, ... | | Authors: | Zhou, K, Hardenbrook, N.J, Liu, S, Cui, Y.X, Krantz, B.A, Zhou, Z.H. | | Deposit date: | 2020-04-13 | | Release date: | 2020-12-16 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Atomic Structures of Anthrax Prechannel Bound with Full-Length Lethal and Edema Factors.
Structure, 28, 2020
|
|
2EA1
 
 | | Crystal structure of Ribonuclease I from Escherichia coli COMPLEXED WITH GUANYLYL-2(PRIME),5(PRIME)-GUANOSINE | | Descriptor: | GUANYLYL-2',5'-PHOSPHOGUANOSINE, Ribonuclease I | | Authors: | Zhou, K, Pan, J, Padmanabhan, S, Lim, R.W, Lim, L.W. | | Deposit date: | 2007-01-29 | | Release date: | 2008-01-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Ribonuclease I from Escherichia Coli Complexed with Guanylyl-2(Prime),5(Prime)-Guanosine at 1.80 Angstroms Resolution
To be Published
|
|
3S9V
 
 | | abietadiene synthase from Abies grandis | | Descriptor: | Abietadiene synthase, chloroplastic | | Authors: | Zhou, K, Hoy, J.A, Mann, F.M, Honzatko, R.B, Peters, R.J. | | Deposit date: | 2011-06-02 | | Release date: | 2012-01-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Insights into diterpene cyclization from structure of bifunctional abietadiene synthase from Abies grandis.
J.Biol.Chem., 287, 2012
|
|
1JNQ
 
 | | LIPOXYGENASE-3 (SOYBEAN) COMPLEX WITH EPIGALLOCATHECHIN (EGC) | | Descriptor: | 2-(3,4,5-TRIHYDROXY-PHENYL)-CHROMAN-3,5,7-TRIOL, FE (II) ION, lipoxygenase-3 | | Authors: | Zhou, K, Skrzypczak-Jankun, E, Jankun, J. | | Deposit date: | 2001-07-24 | | Release date: | 2003-06-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Inhibition of lipoxygenase by (-)-epigallocatechin gallate: X-ray analysis at 2.1 A reveals degradation of EGCG
and shows soybean LOX-3 complex with EGC instead.
INT.J.MOL.MED., 12, 2003
|
|
6UPL
 
 | | Structure of FACT_subnucleosome complex 2 | | Descriptor: | DNA (79-mer), FACT complex subunit SPT16, FACT complex subunit SSRP1, ... | | Authors: | Zhou, K, Tan, Y.Z, Wei, H, Liu, Y, Carragher, B, Potter, C, Luger, K. | | Deposit date: | 2019-10-17 | | Release date: | 2019-12-11 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (7.4 Å) | | Cite: | FACT caught in the act of manipulating the nucleosome.
Nature, 577, 2020
|
|
6UPK
 
 | | Structure of FACT_subnucleosome complex 1 | | Descriptor: | DNA (79-mer), FACT complex subunit SPT16, FACT complex subunit SSRP1, ... | | Authors: | Zhou, K, Tan, Y.Z, Wei, H, Liu, Y, Carragher, B, Potter, C, Luger, K. | | Deposit date: | 2019-10-17 | | Release date: | 2019-12-11 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | FACT caught in the act of manipulating the nucleosome.
Nature, 577, 2020
|
|
4QLA
 
 | | Crystal structure of juvenile hormone epoxide hydrolase from the silkworm Bombyx mori | | Descriptor: | Juvenile hormone epoxide hydrolase, PENTAETHYLENE GLYCOL | | Authors: | Zhou, K, Jia, N, Hu, C, Jiang, Y.L, Yang, J.P, Chen, Y, Li, S, Zhou, C.Z. | | Deposit date: | 2014-06-11 | | Release date: | 2014-09-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of juvenile hormone epoxide hydrolase from the silkworm Bombyx mori.
Proteins, 82, 2014
|
|
7UWS
 
 | | Atomic model of the partial VSV nucleocapsid | | Descriptor: | Matrix protein, Nucleoprotein, RNA | | Authors: | Zhou, K, Si, Z, Ge, P, Tsao, J, Luo, M, Zhou, Z.H. | | Deposit date: | 2022-05-03 | | Release date: | 2022-09-28 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Atomic model of vesicular stomatitis virus and mechanism of assembly.
Nat Commun, 13, 2022
|
|
7UAK
 
 | |
7U46
 
 | |
7U47
 
 | |
7U4D
 
 | |
5WXY
 
 | | Crystal structure of Microcystis aeruginosa PCC 7806 aspartate racemase in complex with L-aspartate | | Descriptor: | ASPARTIC ACID, McyF | | Authors: | Cao, D.D, Zhou, K, Jiang, Y.L, Zhou, C.Z. | | Deposit date: | 2017-01-09 | | Release date: | 2018-01-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Structural insights into the catalysis and substrate specificity of cyanobacterial aspartate racemase McyF.
Biochem.Biophys.Res.Commun., 514, 2019
|
|
5WXX
 
 | | Crystal structure of Microcystis aeruginosa PCC 7806 aspartate racemase in complex with citrate | | Descriptor: | CITRIC ACID, McyF | | Authors: | Cao, D.D, Zhou, K, Jiang, Y.L, Zhou, C.Z. | | Deposit date: | 2017-01-09 | | Release date: | 2018-01-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural insights into the catalysis and substrate specificity of cyanobacterial aspartate racemase McyF.
Biochem.Biophys.Res.Commun., 514, 2019
|
|
5J3R
 
 | | Crystal structure of yeast monothiol glutaredoxin Grx6 in complex with a glutathione-coordinated [2Fe-2S] cluster | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, GLUTATHIONE, Monothiol glutaredoxin-6 | | Authors: | Abdalla, M, Dai, Y.-N, Chi, C.-B, Cheng, W, Cao, D.-D, Zhou, K, Ali, W, Chen, Y, Zhou, C.-Z. | | Deposit date: | 2016-03-31 | | Release date: | 2016-10-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Crystal structure of yeast monothiol glutaredoxin Grx6 in complex with a glutathione-coordinated [2Fe-2S] cluster
Acta Crystallogr.,Sect.F, 72, 2016
|
|
6NSJ
 
 | | CryoEM structure of Helicobacter pylori urea channel in closed state | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHATE, Acid-activated urea channel | | Authors: | Cui, Y.X, Zhou, K, Strugatsky, D, Wen, Y, Sachs, G, Munson, K, Zhou, Z.H. | | Deposit date: | 2019-01-24 | | Release date: | 2019-04-03 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | pH-dependent gating mechanism of theHelicobacter pyloriurea channel revealed by cryo-EM.
Sci Adv, 5, 2019
|
|
6NSK
 
 | | CryoEM structure of Helicobacter pylori urea channel in open state. | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHATE, Acid-activated urea channel | | Authors: | Cui, Y.X, Zhou, K, Strugatsky, D, Wen, Y, Sachs, G, Munson, K, Zhou, Z.H. | | Deposit date: | 2019-01-24 | | Release date: | 2019-04-03 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | pH-dependent gating mechanism of theHelicobacter pyloriurea channel revealed by cryo-EM.
Sci Adv, 5, 2019
|
|
3J8Y
 
 | | High-resolution structure of ATP analog-bound kinesin on microtubules | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Shang, Z, Zhou, K, Xu, C, Csencsits, R, Cochran, J.C, Sindelar, C.V. | | Deposit date: | 2014-11-20 | | Release date: | 2014-12-10 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | High-resolution structures of kinesin on microtubules provide a basis for nucleotide-gated force-generation.
Elife, 3, 2014
|
|
6PSN
 
 | | Anthrax toxin protective antigen channels bound to lethal factor | | Descriptor: | CALCIUM ION, Lethal factor, Protective antigen | | Authors: | Hardenbrook, N.J, Liu, S, Zhou, K, Zhou, Z.H, Krantz, B.A. | | Deposit date: | 2019-07-12 | | Release date: | 2020-03-04 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Atomic structures of anthrax toxin protective antigen channels bound to partially unfolded lethal and edema factors.
Nat Commun, 11, 2020
|
|
4K6N
 
 | | Crystal structure of yeast 4-amino-4-deoxychorismate lyase | | Descriptor: | Aminodeoxychorismate lyase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Dai, Y.-N, Chi, C.-B, Zhou, K, Cheng, W, Jiang, Y.-L, Ren, Y.-M, Chen, Y, Zhou, C.-Z. | | Deposit date: | 2013-04-16 | | Release date: | 2013-07-10 | | Last modified: | 2013-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and catalytic mechanism of yeast 4-amino-4-deoxychorismate lyase
J.Biol.Chem., 288, 2013
|
|
1SJF
 
 | | Crystal Structure of the Hepatitis Delta Virus Gemonic Ribozyme Precursor, with C75U mutaion, in Cobalt Hexammine solution | | Descriptor: | COBALT HEXAMMINE(III), Hepatitis Delta virus ribozyme, U1 small nuclear ribonucleoprotein A | | Authors: | Ke, A, Zhou, K, Ding, F, Cate, J.H.D, Doudna, J.A. | | Deposit date: | 2004-03-03 | | Release date: | 2004-05-18 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | A conformational switch controls hepatitis delta virus ribozyme catalysis.
Nature, 429, 2004
|
|
1SJ4
 
 | | Crystal structure of a C75U mutant Hepatitis Delta Virus ribozyme precursor, in Cu2+ solution | | Descriptor: | precursor form of the Hepatitis Delta virus ribozyme, small nuclear ribonucleoprotein A | | Authors: | Ke, A, Zhou, K, Ding, F, Cate, J.H, Doudna, J.A. | | Deposit date: | 2004-03-02 | | Release date: | 2004-05-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A Conformational Switch controls hepatitis delta virus ribozyme
catalysis
Nature, 429, 2004
|
|
