8HE7
 
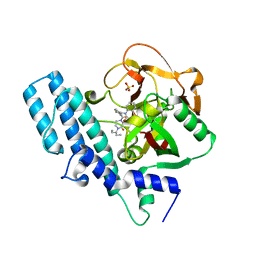 | | ADP-ribosyltransferase 1 (PARP1) catalytic domain bound to a quinazoline-2,4(1H,3H)-dione inhibitor | | Descriptor: | 1-[[4-fluoranyl-3-(3-oxidanylidene-4-pentan-3-yl-piperazin-1-yl)carbonyl-phenyl]methyl]quinazoline-2,4-dione, Poly [ADP-ribose] polymerase 1, processed C-terminus, ... | | Authors: | Wang, X.Y, Zhou, J, Xu, B.L. | | Deposit date: | 2022-11-07 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of Quinazoline-2,4(1 H ,3 H )-dione Derivatives Containing a Piperizinone Moiety as Potent PARP-1/2 Inhibitors─Design, Synthesis, In Vivo Antitumor Activity, and X-ray Crystal Structure Analysis.
J.Med.Chem., 66, 2023
|
|
8HE8
 
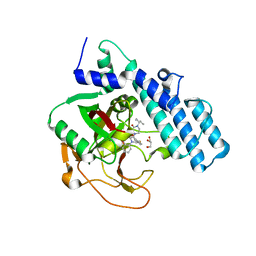 | | Human ADP-ribosyltransferase 2 (PARP2) catalytic domain bound to a quinazoline-2,4(1H,3H)-dione inhibitor | | Descriptor: | 1-[[4-fluoranyl-3-(3-oxidanylidene-4-pentan-3-yl-piperazin-1-yl)carbonyl-phenyl]methyl]quinazoline-2,4-dione, GLYCEROL, Poly [ADP-ribose] polymerase 2 | | Authors: | Wang, X.Y, Xu, B.L, Zhou, J. | | Deposit date: | 2022-11-07 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Discovery of Quinazoline-2,4(1 H ,3 H )-dione Derivatives Containing a Piperizinone Moiety as Potent PARP-1/2 Inhibitors─Design, Synthesis, In Vivo Antitumor Activity, and X-ray Crystal Structure Analysis.
J.Med.Chem., 66, 2023
|
|
7VZU
 
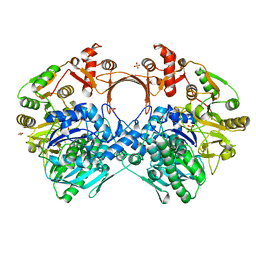 | | The structure of GdmN Y82F mutant | | Descriptor: | 1,2-ETHANEDIOL, FE (III) ION, GdmN, ... | | Authors: | Wei, J, Zheng, J, Zhou, J, Kang, Q, Bai, L. | | Deposit date: | 2021-11-16 | | Release date: | 2022-11-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Endowing homodimeric carbamoyltransferase GdmN with iterative functions through structural characterization and mechanistic studies.
Nat Commun, 13, 2022
|
|
7VX0
 
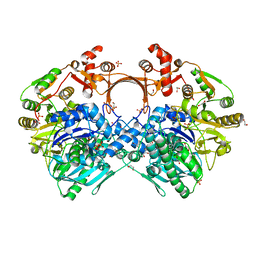 | | The structure of GdmN complex with ATP | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, FE (III) ION, ... | | Authors: | Wei, J, Zheng, J, Zhou, J, Kang, Q, Bai, L. | | Deposit date: | 2021-11-12 | | Release date: | 2022-11-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Endowing homodimeric carbamoyltransferase GdmN with iterative functions through structural characterization and mechanistic studies.
Nat Commun, 13, 2022
|
|
7VYO
 
 | | The structure of GdmN | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Wei, J, Zheng, J, Zhou, J, Kang, Q, Bai, L. | | Deposit date: | 2021-11-14 | | Release date: | 2022-11-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Endowing homodimeric carbamoyltransferase GdmN with iterative functions through structural characterization and mechanistic studies.
Nat Commun, 13, 2022
|
|
7VZQ
 
 | | The structure of GdmN V24Y/G157A/R158A/G188R mutant in complex with carbamoyl adenylate intermediate | | Descriptor: | 1,2-ETHANEDIOL, 5'-O-[(S)-(carbamoyloxy)(hydroxy)phosphoryl]adenosine, CHLORIDE ION, ... | | Authors: | Wei, J, Zheng, J, Zhou, J, Kang, Q, Bai, L. | | Deposit date: | 2021-11-16 | | Release date: | 2022-11-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Endowing homodimeric carbamoyltransferase GdmN with iterative functions through structural characterization and mechanistic studies.
Nat Commun, 13, 2022
|
|
7VYJ
 
 | | The structure of GdmN in complex with carbamoyl adenylate intermediate | | Descriptor: | 1,2-ETHANEDIOL, 5'-O-[(S)-(carbamoyloxy)(hydroxy)phosphoryl]adenosine, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Wei, J, Zheng, J, Zhou, J, Kang, Q, Bai, L. | | Deposit date: | 2021-11-14 | | Release date: | 2022-11-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Endowing homodimeric carbamoyltransferase GdmN with iterative functions through structural characterization and mechanistic studies.
Nat Commun, 13, 2022
|
|
7VZZ
 
 | | The structure of GdmN in complex with the natural tetrahedral intermediate, carbamoyl adenylate, and 20-O-methyl-19-chloroproansamitocin | | Descriptor: | (5~{S},6~{E},8~{S},9~{S},12~{R},15~{E})-21-chloranyl-12,20-dimethoxy-6,8,16-trimethyl-5,9-bis(oxidanyl)-2-azabicyclo[16.3.1]docosa-1(21),6,15,18(22),19-pentaene-3,11-dione, 1,2-ETHANEDIOL, 5'-O-[(S)-(carbamoyloxy)(hydroxy)phosphoryl]adenosine, ... | | Authors: | Wei, J, Zheng, J, Zhou, J, Kang, Q, Bai, L. | | Deposit date: | 2021-11-17 | | Release date: | 2022-11-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Endowing homodimeric carbamoyltransferase GdmN with iterative functions through structural characterization and mechanistic studies.
Nat Commun, 13, 2022
|
|
7VYP
 
 | | The structure of GdmN complex with the natural tetrahedral intermediate, carbamoylated derivative, and AMP | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE MONOPHOSPHATE, FE (III) ION, ... | | Authors: | Wei, J, Zheng, J, Zhou, J, Kang, Q, Bai, L. | | Deposit date: | 2021-11-15 | | Release date: | 2022-11-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Endowing homodimeric carbamoyltransferase GdmN with iterative functions through structural characterization and mechanistic studies.
Nat Commun, 13, 2022
|
|
7VZN
 
 | | The structure of GdmN in complex with carbamoyl adenylate intermediate and 20-O-methyl-19-chloroproansamitocin | | Descriptor: | (5~{S},6~{E},8~{S},9~{S},12~{R},15~{E})-21-chloranyl-12,20-dimethoxy-6,8,16-trimethyl-5,9-bis(oxidanyl)-2-azabicyclo[16.3.1]docosa-1(21),6,15,18(22),19-pentaene-3,11-dione, 1,2-ETHANEDIOL, 5'-O-[(S)-(carbamoyloxy)(hydroxy)phosphoryl]adenosine, ... | | Authors: | Wei, J, Zheng, J, Zhou, J, Kang, Q, Bai, L. | | Deposit date: | 2021-11-16 | | Release date: | 2022-11-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Endowing homodimeric carbamoyltransferase GdmN with iterative functions through structural characterization and mechanistic studies.
Nat Commun, 13, 2022
|
|
7VZY
 
 | | The structure of GdmN complex with AMP and 20-O-methyl-19-chloroproansamitocin | | Descriptor: | (5~{S},6~{E},8~{S},9~{S},12~{R},15~{E})-21-chloranyl-12,20-dimethoxy-6,8,16-trimethyl-5,9-bis(oxidanyl)-2-azabicyclo[16.3.1]docosa-1(21),6,15,18(22),19-pentaene-3,11-dione, 1,2-ETHANEDIOL, ADENOSINE MONOPHOSPHATE, ... | | Authors: | Wei, J, Zheng, J, Zhou, J, Kang, Q, Bai, L. | | Deposit date: | 2021-11-17 | | Release date: | 2022-11-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Endowing homodimeric carbamoyltransferase GdmN with iterative functions through structural characterization and mechanistic studies.
Nat Commun, 13, 2022
|
|
4RNC
 
 | | Crystal structure of an esterase RhEst1 from Rhodococcus sp. ECU1013 | | Descriptor: | Esterase, PHOSPHATE ION | | Authors: | Dou, S, Kong, X.D, Xu, J.H, Zhou, J. | | Deposit date: | 2014-10-23 | | Release date: | 2015-10-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Substrate channel evolution of an esterase for the synthesis of Cilastatin
CATALYSIS SCIENCE AND TECHNOLOGY, 5, 2015
|
|
5X5R
 
 | |
7WVS
 
 | | The structure of FinI complex with SAM | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Methyltransf_2 domain-containing protein, ... | | Authors: | Lu, J, Zhou, J. | | Deposit date: | 2022-02-11 | | Release date: | 2023-02-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The structure of FinI complex with SAM
To Be Published
|
|
8HKV
 
 | |
8HKX
 
 | |
8HKU
 
 | |
8HKY
 
 | |
8HL5
 
 | |
8HL4
 
 | |
8JLV
 
 | |
8JNX
 
 | |
8JN0
 
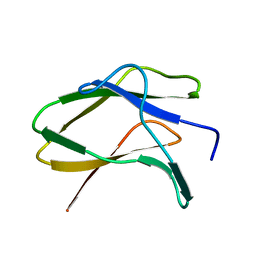 | | N/F domain of alkaline amylase Amy703 | | Descriptor: | Alpha-amylase | | Authors: | Xiang, L, Zhang, G, Zhou, J. | | Deposit date: | 2023-06-05 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.22826374 Å) | | Cite: | Truncation of N-terminus domain of alkaline a-amylase to form a unique dimer leads to improved activity and stability and decreased calcium ion dependence
To Be Published
|
|
5HSJ
 
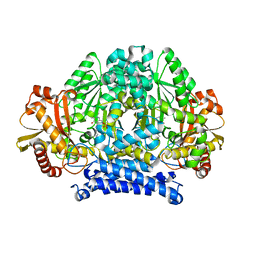 | | Structure of tyrosine decarboxylase complex with PLP at 1.9 Angstroms resolution | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, Putative decarboxylase | | Authors: | Ni, Y, Zhou, J, Zhu, H, Zhang, K. | | Deposit date: | 2016-01-25 | | Release date: | 2016-09-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of tyrosine decarboxylase and identification of key residues involved in conformational swing and substrate binding
Sci Rep, 6, 2016
|
|
7VDO
 
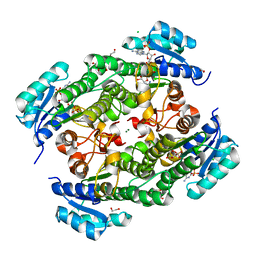 | | Crystal structure of KRED F147L/L153Q/Y190P variant | | Descriptor: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-alpha-(Or 20-beta)-hydroxysteroid dehydrogenase, ... | | Authors: | Cui, J, Huang, X, Wang, B, Zhao, H, Zhou, J. | | Deposit date: | 2021-09-07 | | Release date: | 2022-05-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85571563 Å) | | Cite: | Photoinduced chemomimetic biocatalysis for enantioselective intermolecular radical conjugate addition
Nat Catal, 2022
|
|
