7V2T
 
 | | The complex structure of SoBcmB and its natural precursor 2 | | Descriptor: | (3S,6S)-3-((R)-2,3-dihydroxy-2-methylpropyl)-6-((S)-4-hydroxybutan-2-yl)piperazine-2,5-dione, 2-OXOGLUTARIC ACID, CHLORIDE ION, ... | | Authors: | Zhou, J.H, Wu, L. | | Deposit date: | 2021-08-09 | | Release date: | 2023-02-15 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.20005679 Å) | | Cite: | Enzymatic catalysis favours eight-membered over five-membered ring closure in bicyclomycin biosynthesis
Nat Catal, 6, 2023
|
|
3LWP
 
 | | Structure of H/ACA RNP bound to a substrate RNA containing 5BrdU | | Descriptor: | 5'-R(*GP*AP*GP*CP*GP*(BRU)P*GP*CP*GP*GP*UP*UP*U)-3, 50S ribosomal protein L7Ae, H/ACA RNA, ... | | Authors: | Zhou, J, Liang, B, Lv, C, Yang, W, Li, H. | | Deposit date: | 2010-02-24 | | Release date: | 2010-07-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Glycosidic bond conformation preference plays a pivotal role in catalysis of RNA pseudouridylation: a combined simulation and structural study.
J.Mol.Biol., 401, 2010
|
|
8FS1
 
 | | CamA Adenine Methyltransferase Complexed to Cognate Substrate DNA and Inhibitor 11a (YD905) | | Descriptor: | 1,2-ETHANEDIOL, 5'-S-{2-[N'-(cyclohexylmethyl)carbamimidamido]ethyl}-N-(3-phenylpropyl)-5'-thioadenosine, DNA (5'-D(*AP*TP*GP*GP*GP*AP*CP*TP*TP*TP*TP*TP*GP*A)-3'), ... | | Authors: | Zhou, J, Horton, J.R, Cheng, X. | | Deposit date: | 2023-01-09 | | Release date: | 2023-05-10 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Comparative Study of Adenosine Analogs as Inhibitors of Protein Arginine Methyltransferases and a Clostridioides difficile- Specific DNA Adenine Methyltransferase.
Acs Chem.Biol., 18, 2023
|
|
8FS2
 
 | | CamA Adenine Methyltransferase Complexed to Cognate Substrate DNA and Inhibitor 11b (YD907) | | Descriptor: | 1,2-ETHANEDIOL, 5'-S-{3-[N'-(cyclohexylmethyl)carbamimidamido]propyl}-N-(3-phenylpropyl)-5'-thioadenosine, DNA (5'-D(*TP*TP*CP*AP*AP*AP*AP*AP*GP*TP*CP*CP*CP*A)-3'), ... | | Authors: | Zhou, J, Horton, J.R, Cheng, X. | | Deposit date: | 2023-01-09 | | Release date: | 2023-05-10 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Comparative Study of Adenosine Analogs as Inhibitors of Protein Arginine Methyltransferases and a Clostridioides difficile- Specific DNA Adenine Methyltransferase.
Acs Chem.Biol., 18, 2023
|
|
8HS0
 
 | | The mutant structure of DHAD V178W | | Descriptor: | Dihydroxy-acid dehydratase, chloroplastic, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Zhou, J, Zang, X, Tang, Y, Yan, Y. | | Deposit date: | 2022-12-16 | | Release date: | 2024-01-17 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Structural bases of dihydroxy acid dehydratase inhibition and biodesign for self-resistance
Biodes Res, 0, 2024
|
|
8IDZ
 
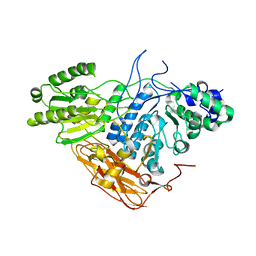 | |
7ETK
 
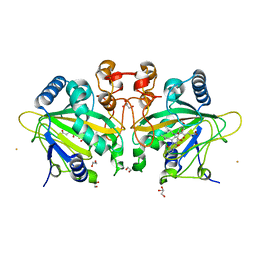 | |
7ETL
 
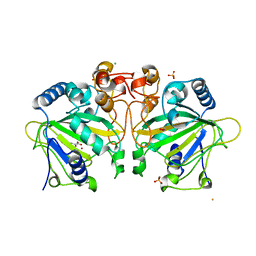 | | The crystal structure of FtmOx1-Y68F | | Descriptor: | 1,2-ETHANEDIOL, 2-OXOGLUTARIC ACID, CHLORIDE ION, ... | | Authors: | Zhou, J.H, Wu, L. | | Deposit date: | 2021-05-13 | | Release date: | 2021-12-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.992128 Å) | | Cite: | Structural Insight into the Catalytic Mechanism of the Endoperoxide Synthase FtmOx1.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
8IKZ
 
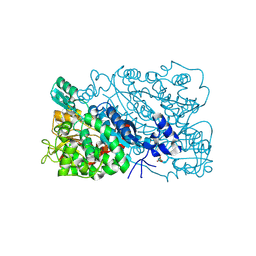 | | The mutant structure of DHAD | | Descriptor: | Dihydroxy-acid dehydratase, chloroplastic, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Zhou, J, Zang, X, Tang, Y, Yan, Y. | | Deposit date: | 2023-03-01 | | Release date: | 2024-03-06 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural bases of dihydroxy acid dehydratase inhibition and biodesign for self-resistance
Biodes Res, 0, 2024
|
|
8I12
 
 | | InuAMN8 | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Glycosyl hydrolase family 32 exo-inulinase | | Authors: | Zhou, J.P, Cen, X.L, He, L.M, Zhang, R, Huang, Z.X. | | Deposit date: | 2023-01-12 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Cold-active and NaCl-tolerant exo-inulinase InuAMN8.
To Be Published
|
|
6KCT
 
 | |
6KBF
 
 | | Crystal structure of plasmodium lysyl-tRNA synthetase in complex with a cladosporin derivative 3 | | Descriptor: | (3~{S})-3-[[(1~{S},3~{S})-3-methylcyclohexyl]methyl]-6,8-bis(oxidanyl)-3,4-dihydro-2~{H}-isoquinolin-1-one, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Zhou, J, Fang, P. | | Deposit date: | 2019-06-24 | | Release date: | 2020-04-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Atomic Resolution Analyses of Isocoumarin Derivatives for Inhibition of Lysyl-tRNA Synthetase.
Acs Chem.Biol., 15, 2020
|
|
8JF9
 
 | |
5CZW
 
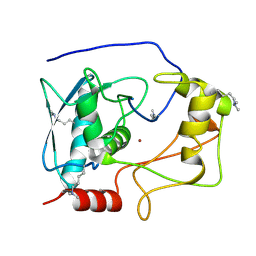 | | Crystal structure of myroilysin | | Descriptor: | Myroilysin, ZINC ION | | Authors: | Zhou, J, Ran, T, Xu, D, Wang, W. | | Deposit date: | 2015-08-01 | | Release date: | 2016-08-03 | | Last modified: | 2017-04-26 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Myroilysin is a New Bacterial Member of the M12A Family of Metzincin Metallopeptidases and Activated by a Cysteine-switch Mechanism
J. Biol. Chem., 292, 2017
|
|
8S9N
 
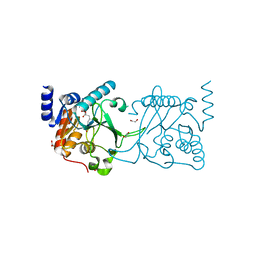 | | DNA cytosine-N4 methyltransferase (residues 61-324) from the Bdelloid rotifer Adineta vaga - C2 crystal form | | Descriptor: | 1,2-ETHANEDIOL, DNA cytosine-N4 methyltransferase, SINEFUNGIN | | Authors: | Zhou, J, Horton, J.R, Cheng, X. | | Deposit date: | 2023-03-29 | | Release date: | 2023-08-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Biochemical and structural characterization of the first-discovered metazoan DNA cytosine-N4 methyltransferase from the bdelloid rotifer Adineta vaga.
J.Biol.Chem., 299, 2023
|
|
2IHX
 
 | |
8I0X
 
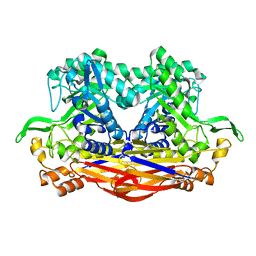 | | Beta-Xylosidase JB13GH39P28 showing salt/ethanol/trypsin tolerance, low-pH/low-Temperature activity, and transformation of notoginsenosides | | Descriptor: | Glycoside hydrolase family 39 beta-xylosidase | | Authors: | Zhou, J.P, Cao, L.J, Lin, M.Y, Zhang, R, Huang, Z.X. | | Deposit date: | 2023-01-11 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | beta-Xylosidase JB13GH39P28(D41G)showing salt/ethanol/trypsin tolerance and transformation of notoginsenosides
To Be Published
|
|
8S9M
 
 | | DNA cytosine-N4 methyltransferase (residues 79-324) from the Bdelloid rotifer Adineta vaga | | Descriptor: | 1,2-ETHANEDIOL, DNA cytosine-N4 methyltransferase, SINEFUNGIN | | Authors: | Zhou, J, Horton, J.R, Cheng, X. | | Deposit date: | 2023-03-29 | | Release date: | 2023-08-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Biochemical and structural characterization of the first-discovered metazoan DNA cytosine-N4 methyltransferase from the bdelloid rotifer Adineta vaga.
J.Biol.Chem., 299, 2023
|
|
8S9O
 
 | | DNA cytosine-N4 methyltransferase (residues 61-324) from the Bdelloid rotifer Adineta vaga - P1 crystal form | | Descriptor: | 1,2-ETHANEDIOL, DNA cytosine-N4 methyltransferase, SINEFUNGIN | | Authors: | Zhou, J, Horton, J.R, Cheng, X. | | Deposit date: | 2023-03-29 | | Release date: | 2023-08-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Biochemical and structural characterization of the first-discovered metazoan DNA cytosine-N4 methyltransferase from the bdelloid rotifer Adineta vaga.
J.Biol.Chem., 299, 2023
|
|
1OZB
 
 | | Crystal Structure of SecB complexed with SecA C-terminus | | Descriptor: | Preprotein translocase secA subunit, Protein-export protein secB, ZINC ION | | Authors: | Zhou, J, Xu, Z. | | Deposit date: | 2003-04-08 | | Release date: | 2003-10-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural determinants of SecB recognition by SecA in bacterial protein translocation
NAT.STRUCT.BIOL., 10, 2003
|
|
8CY5
 
 | | CamA Adenine Methyltransferase Complexed to Cognate Substrate DNA and Compound 39 | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*TP*TP*CP*AP*AP*AP*AP*AP*GP*TP*CP*CP*CP*A)-3'), N-{3-[1-(tert-butoxycarbonyl)piperidin-4-yl]propyl}adenosine, ... | | Authors: | Zhou, J, Horton, J.R, Cheng, X. | | Deposit date: | 2022-05-22 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Systematic Design of Adenosine Analogs as Inhibitors of a Clostridioides difficile- Specific DNA Adenine Methyltransferase Required for Normal Sporulation and Persistence.
J.Med.Chem., 66, 2023
|
|
8CY3
 
 | | CamA Adenine Methyltransferase Complexed to Cognate Substrate DNA and Compound 15 | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*TP*TP*CP*AP*AP*AP*AP*AP*GP*TP*CP*CP*CP*A)-3'), N-[3-(4-aminophenyl)propyl]adenosine, ... | | Authors: | Zhou, J, Horton, J.R, Cheng, X. | | Deposit date: | 2022-05-22 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Systematic Design of Adenosine Analogs as Inhibitors of a Clostridioides difficile- Specific DNA Adenine Methyltransferase Required for Normal Sporulation and Persistence.
J.Med.Chem., 66, 2023
|
|
8IMU
 
 | | Dihydroxyacid dehydratase (DHAD) mutant-V497F | | Descriptor: | ACETATE ION, Dihydroxy-acid dehydratase, chloroplastic, ... | | Authors: | Zhou, J, Zang, X, Tang, Y, Yan, Y. | | Deposit date: | 2023-03-07 | | Release date: | 2024-03-13 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural bases of dihydroxy acid dehydratase inhibition and biodesign for self-resistance
Biodes Res, 0, 2024
|
|
8CY1
 
 | | CamA Adenine Methyltransferase Complexed to Cognate Substrate DNA and Compound 19 | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*AP*TP*GP*GP*GP*AP*CP*TP*TP*TP*TP*TP*GP*A)-3'), N-(5-phenylpentyl)adenosine, ... | | Authors: | Zhou, J, Horton, J.R, Cheng, X. | | Deposit date: | 2022-05-22 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Systematic Design of Adenosine Analogs as Inhibitors of a Clostridioides difficile- Specific DNA Adenine Methyltransferase Required for Normal Sporulation and Persistence.
J.Med.Chem., 66, 2023
|
|
8CY4
 
 | | CamA Adenine Methyltransferase Complexed to Cognate Substrate DNA and Compound 16 | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*AP*TP*GP*GP*GP*AP*CP*TP*TP*TP*TP*TP*GP*A)-3'), N-[3-(4-hydroxyphenyl)propyl]adenosine, ... | | Authors: | Zhou, J, Horton, J.R, Cheng, X. | | Deposit date: | 2022-05-22 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Systematic Design of Adenosine Analogs as Inhibitors of a Clostridioides difficile- Specific DNA Adenine Methyltransferase Required for Normal Sporulation and Persistence.
J.Med.Chem., 66, 2023
|
|
