7VZB
 
 | | Cryo-EM structure of C22:0-CoA bound human very long-chain fatty acid ABC transporter ABCD1 | | Descriptor: | CHOLESTEROL HEMISUCCINATE, Peroxisomal Membrane Protein related,ATP-binding cassette sub-family D member 1, S-[2-[3-[[(2R)-4-[[[(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-4-oxidanyl-3-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxy-3,3-dimethyl-2-oxidanyl-butanoyl]amino]propanoylamino]ethyl] docosanethioate | | Authors: | Chen, Z.P, Xu, D, Wang, L, Mao, Y.X, Yang, L, Cheng, M.T, Hou, W.T, Chen, Y.X, Zhou, C.Z. | | Deposit date: | 2021-11-15 | | Release date: | 2022-05-18 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.59 Å) | | Cite: | Structural basis of substrate recognition and translocation by human very long-chain fatty acid transporter ABCD1.
Nat Commun, 13, 2022
|
|
5XNJ
 
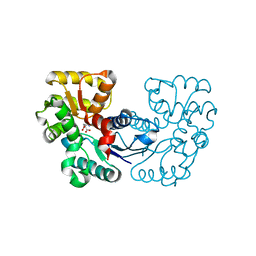 | |
5XNK
 
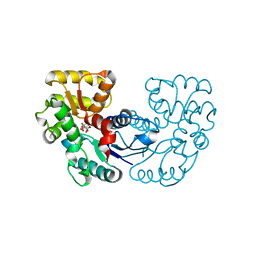 | | Crystal structure of Microcystis aeruginosa PCC 7806 aspartate racemase in complex with DL-methyl-aspartate | | Descriptor: | (2S,3S)-3-methyl-aspartic acid, 3-METHYL-BETA-D-ASPARTIC ACID, McyF | | Authors: | Cao, D.D, Zhou, K, Jiang, Y.L, Zhou, C.Z. | | Deposit date: | 2017-05-23 | | Release date: | 2018-05-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structure-function Analyses of a Cyanobacterial Aspartate Racemase Reveal Its Catalytic Mechanism and Substrate Specificity
To Be Published
|
|
5XNI
 
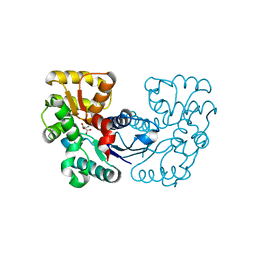 | |
8HDS
 
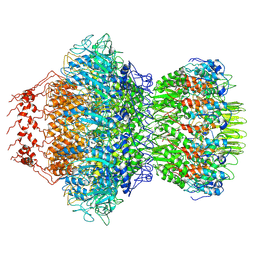 | | Cyanophage Pam3 portal-adaptor | | Descriptor: | Pam3 adaptor protein, Pam3 portal protein | | Authors: | Yang, F, Jiang, Y.L, Zhou, C.Z. | | Deposit date: | 2022-11-06 | | Release date: | 2023-01-18 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | Fine structure and assembly pattern of a minimal myophage Pam3.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8HDT
 
 | |
8HDW
 
 | | Cyanophage Pam3 Sheath-tube | | Descriptor: | Pam3 sheath protein, pam3 tube | | Authors: | Yang, F, Jiang, Y.L, Zhou, C.Z. | | Deposit date: | 2022-11-06 | | Release date: | 2023-01-18 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Fine structure and assembly pattern of a minimal myophage Pam3.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8HDR
 
 | | Cyanophage Pam3 neck | | Descriptor: | Pam3 adaptor protein, Pam3 connector protein, Pam3 sheath protein, ... | | Authors: | Yang, F, Jiang, Y.L, Zhou, C.Z. | | Deposit date: | 2022-11-06 | | Release date: | 2023-02-01 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.66 Å) | | Cite: | Fine structure and assembly pattern of a minimal myophage Pam3.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8I4B
 
 | | Cryo-EM structure of apo-form ABCC4 | | Descriptor: | ATP-binding cassette sub-family C member 4 | | Authors: | Chen, Y, Wang, L, Hou, W.T, Zhou, C.Z, Chen, Y, Li, Q. | | Deposit date: | 2023-01-19 | | Release date: | 2023-05-24 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Cryo-EM structure ofABCC4
Nat Cardiovasc Res, 2023
|
|
8I4A
 
 | | Cryo-EM structure of dipyridamole-bound ABCC4 | | Descriptor: | 2-[[2-[bis(2-hydroxyethyl)amino]-4,8-di(piperidin-1-yl)pyrimido[5,4-d]pyrimidin-6-yl]-(2-hydroxyethyl)amino]ethanol, ATP-binding cassette sub-family C member 4 | | Authors: | Chen, Y, Wang, L, Hou, W.T, Zhou, C.Z, Chen, Y, Li, Q. | | Deposit date: | 2023-01-19 | | Release date: | 2023-05-24 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structure ofABCC4
Nat Cardiovasc Res, 2023
|
|
8I4C
 
 | | Cryo-EM structure of U46619-bound ABCC4 | | Descriptor: | (5Z)-7-{(1R,4S,5S,6R)-6-[(1E,3S)-3-hydroxyoct-1-en-1-yl]-2-oxabicyclo[2.2.1]hept-5-yl}hept-5-enoic acid, ATP-binding cassette sub-family C member 4 | | Authors: | Chen, Y, Wang, L, Hou, W.T, Zhou, C.Z, Chen, Y, Li, Q. | | Deposit date: | 2023-01-19 | | Release date: | 2023-05-24 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Cryo-EM structure ofABCC4
Nat Cardiovasc Res, 2023
|
|
8HW4
 
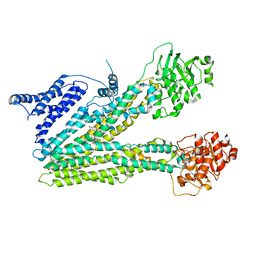 | | Cryo-EM structure of dehydroepiandrosterone sulfate-bound human ABC transporter ABCC3 in nanodiscs | | Descriptor: | 17-oxoandrost-5-en-3beta-yl hydrogen sulfate, ATP-binding cassette sub-family C member 3 | | Authors: | Wang, J, Wang, F.F, Chen, Y.X, Zhou, C.Z. | | Deposit date: | 2022-12-28 | | Release date: | 2023-06-07 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Placing steroid hormones within the human ABCC3 transporter reveals a compatible amphiphilic substrate-binding pocket.
Embo J., 42, 2023
|
|
8HVH
 
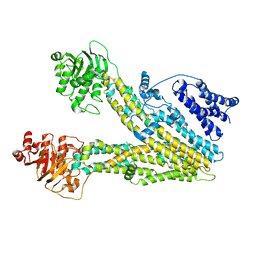 | | Cryo-EM structure of ABC transporter ABCC3 | | Descriptor: | ATP-binding cassette sub-family C member 3 | | Authors: | Wang, J, Wang, F.F, Chen, Y.X, Zhou, C.Z. | | Deposit date: | 2022-12-26 | | Release date: | 2023-06-07 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Placing steroid hormones within the human ABCC3 transporter reveals a compatible amphiphilic substrate-binding pocket.
Embo J., 42, 2023
|
|
8HW2
 
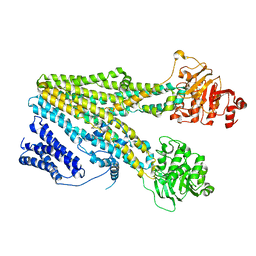 | | Cryo-EM structure of beta-estradiol 17-(beta-D-glucuronide)-bound human ABC transporter ABCC3 in nanodiscs | | Descriptor: | ATP-binding cassette sub-family C member 3, Estradiol-17beta-glucuronide | | Authors: | Wang, J, Wang, F.F, Chen, Y.X, Zhou, C.Z. | | Deposit date: | 2022-12-28 | | Release date: | 2023-06-07 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | Placing steroid hormones within the human ABCC3 transporter reveals a compatible amphiphilic substrate-binding pocket.
Embo J., 42, 2023
|
|
7FDV
 
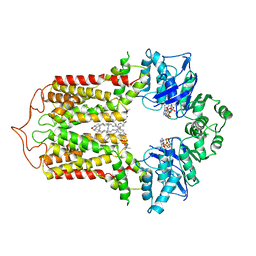 | | Cryo-EM structure of the human cholesterol transporter ABCG1 in complex with cholesterol | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATP-binding cassette sub-family G member 1, CHOLESTEROL, ... | | Authors: | Xu, D, Li, Y.Y, Yang, F.R, Sun, C.R, Pan, J.H, Wang, L, Chen, Z.P, Fang, S.C, Yao, X.B, Hou, W.T, Zhou, C.Z, Chen, Y. | | Deposit date: | 2021-07-18 | | Release date: | 2022-06-22 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Structure and transport mechanism of the human cholesterol transporter ABCG1.
Cell Rep, 38, 2022
|
|
5XU0
 
 | | Structure of the membrane fusion protein Spr0693 from Streptococcus pneumoniae R6 | | Descriptor: | Membrane-fusion protein | | Authors: | Yang, H.B, Jiang, Y.L, Hou, W.T, Chen, M.T, Chen, Y, Zhou, C.Z. | | Deposit date: | 2017-06-22 | | Release date: | 2018-01-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structure of a MacAB-like efflux pump from Streptococcus pneumoniae.
Nat Commun, 9, 2018
|
|
5XU1
 
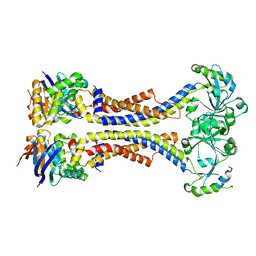 | | Structure of a non-canonical ABC transporter from Streptococcus pneumoniae R6 | | Descriptor: | ABC transporter ATP-binding protein, ABC transporter permeae, MAGNESIUM ION | | Authors: | Yang, H.B, Jiang, Y.L, Hou, W.T, Chen, M.T, Chen, Y, Zhou, C.Z. | | Deposit date: | 2017-06-22 | | Release date: | 2018-01-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of a MacAB-like efflux pump from Streptococcus pneumoniae.
Nat Commun, 9, 2018
|
|
5Y2V
 
 | | Strcutrue of the full-length CcmR complexed with 2-OG from Synechocystis PCC6803 | | Descriptor: | 2-OXOGLUTARIC ACID, PHOSPHATE ION, Rubisco operon transcriptional regulator | | Authors: | Jiang, Y.L, Wang, X.P, Sun, H, Cheng, W, Han, S.J, Li, W.F, Chen, Y, Zhou, C.Z. | | Deposit date: | 2017-07-27 | | Release date: | 2017-12-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Coordinating carbon and nitrogen metabolic signaling through the cyanobacterial global repressor NdhR.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5Y4U
 
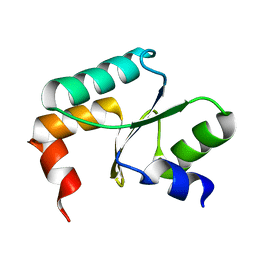 | | Crystal structure of Grx domain of Grx3 from Saccharomyces cerevisiae | | Descriptor: | Monothiol glutaredoxin-3 | | Authors: | Chi, C.B, Tang, Y.J, Zhang, J.H, Dai, Y.N, Abdalla, M, Chen, Y.X, Zhou, C.Z. | | Deposit date: | 2017-08-05 | | Release date: | 2018-08-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and Biochemical Insights into the Multiple Functions of Yeast Grx3.
J.Mol.Biol., 430, 2018
|
|
5Y2W
 
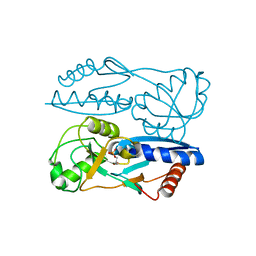 | | Structure of Synechocystis PCC6803 CcmR regulatory domain in complex with 2-PG | | Descriptor: | 2-PHOSPHOGLYCOLIC ACID, Rubisco operon transcriptional regulator | | Authors: | Jiang, Y.L, Wang, X.P, Sun, H, Cheng, W, Cao, D.D, Han, S.J, Li, W.F, Chen, Y, Zhou, C.Z. | | Deposit date: | 2017-07-27 | | Release date: | 2017-12-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Coordinating carbon and nitrogen metabolic signaling through the cyanobacterial global repressor NdhR.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5Y4T
 
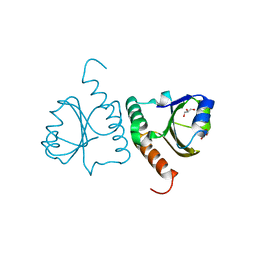 | | Crystal structure of Trx domain of Grx3 from Saccharomyces cerevisiae | | Descriptor: | GLYCEROL, Glutaredoxin | | Authors: | Chi, C.B, Tang, Y.J, Zhang, J.H, Dai, Y.N, Abdalla, M, Chen, Y.X, Zhou, C.Z. | | Deposit date: | 2017-08-05 | | Release date: | 2018-08-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural and Biochemical Insights into the Multiple Functions of Yeast Grx3.
J.Mol.Biol., 430, 2018
|
|
5YRP
 
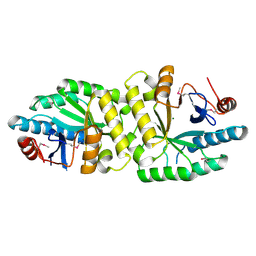 | | Crystal structure of the EAL domain of Mycobacterium smegmatis DcpA | | Descriptor: | MAGNESIUM ION, Sensory box/response regulator | | Authors: | Chen, H.J, li, N, Luo, Y, Jiang, Y.L, Zhou, C.Z, Chen, Y, Li, Q. | | Deposit date: | 2017-11-09 | | Release date: | 2018-05-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | The GDP-switched GAF domain of DcpA modulates the concerted synthesis/hydrolysis of c-di-GMP inMycobacterium smegmatis.
Biochem. J., 475, 2018
|
|
5Y4B
 
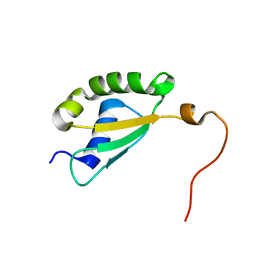 | | Solution structure of yeast Fra2 | | Descriptor: | BolA-like protein 2 | | Authors: | Tang, Y.J, Chi, C.B, Zhang, J.H, Dai, Y.N, Abdalla, M, Chen, Y.X, Zhou, C.Z. | | Deposit date: | 2017-08-03 | | Release date: | 2018-03-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural and Biochemical Insights into the Multiple Functions of Yeast Grx3.
J. Mol. Biol., 430, 2018
|
|
5Z74
 
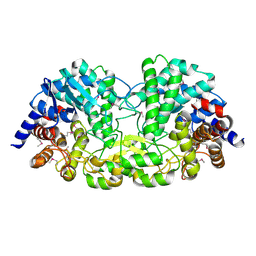 | | Crystal structure of alkaline/neutral invertase InvB from Anabaena sp. PCC 7120 complexed with sucrose | | Descriptor: | Alr0819 protein, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose | | Authors: | Xie, J, Hu, H.X, Cai, K, Yang, F, Jiang, Y.L, Chen, Y, Zhou, C.Z. | | Deposit date: | 2018-01-27 | | Release date: | 2018-05-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural and enzymatic analyses of Anabaena heterocyst-specific alkaline invertase InvB.
FEBS Lett., 592, 2018
|
|
5Z73
 
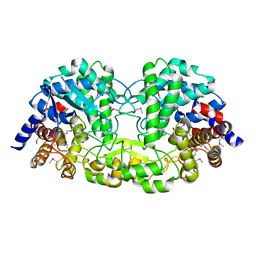 | | Crystal structure of alkaline/neutral invertase InvB from Anabaena sp. PCC 7120 | | Descriptor: | Alr0819 protein, GLYCEROL | | Authors: | Xie, J, Hu, H.X, Cai, K, Yang, F, Jiang, Y.L, Chen, Y, Zhou, C.Z. | | Deposit date: | 2018-01-27 | | Release date: | 2018-05-30 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural and enzymatic analyses of Anabaena heterocyst-specific alkaline invertase InvB.
FEBS Lett., 592, 2018
|
|
