8WM8
 
 | | Cryo-EM structure of cyanobacterial nitrate/nitrite transporter NrtBCD in complex with nitrate | | Descriptor: | NITRATE ION, Nitrate transport ATP-binding protein, Nitrate transport permease protein | | Authors: | Li, B, Zhou, C.Z, Chen, Y.X, Jiang, Y.L. | | Deposit date: | 2023-10-03 | | Release date: | 2024-02-28 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Allosteric regulation of nitrate transporter NRT via the signaling protein PII.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8WM7
 
 | | Cryo-EM structure of cyanobacterial nitrate/nitrite transporter NrtBCD in complex with signalling protein PII | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Nitrate transport ATP-binding protein, Nitrate transport permease protein, ... | | Authors: | Li, B, Zhou, C.Z, Chen, Y.X, Jiang, Y.L. | | Deposit date: | 2023-10-03 | | Release date: | 2024-02-28 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | Allosteric regulation of nitrate transporter NRT via the signaling protein PII.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
5GOR
 
 | | Crystal structure of alkaline invertase InvA from Anabaena sp. PCC 7120 | | Descriptor: | Alkaline Invertase, GLYCEROL, SULFATE ION | | Authors: | Xie, J, Cai, K, Hu, H.X, Jiang, Y.L, Yang, F, Hu, P.F, Chen, Y, Zhou, C.Z. | | Deposit date: | 2016-07-28 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.673 Å) | | Cite: | Structural Analysis of the Catalytic Mechanism and Substrate Specificity of Anabaena Alkaline Invertase InvA Reveals a Novel Glucosidase
J. Biol. Chem., 291, 2016
|
|
5GOP
 
 | | Crystal structure of alkaline invertase InvA from Anabaena sp. PCC 7120 complexed with sucrose | | Descriptor: | Alkaline Invertase, beta-D-fructofuranose, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose | | Authors: | Xie, J, Cai, K, Hu, H.X, Jiang, Y.L, Yang, F, Hu, P.F, Chen, Y, Zhou, C.Z. | | Deposit date: | 2016-07-28 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural Analysis of the Catalytic Mechanism and Substrate Specificity of Anabaena Alkaline Invertase InvA Reveals a Novel Glucosidase
J. Biol. Chem., 291, 2016
|
|
5GVV
 
 | | Crystal structure of the glycosyltransferase GlyE in Streptococcus pneumoniae TIGR4 | | Descriptor: | Glycosyl transferase family 8, MANGANESE (II) ION, URIDINE-5'-DIPHOSPHATE | | Authors: | Jiang, Y.L, Jin, H, Zhao, R.L, Yang, H.B, Chen, Y, Zhou, C.Z. | | Deposit date: | 2016-09-07 | | Release date: | 2017-03-01 | | Last modified: | 2017-12-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Defining the enzymatic pathway for polymorphic O-glycosylation of the pneumococcal serine-rich repeat protein PsrP.
J. Biol. Chem., 292, 2017
|
|
5GOQ
 
 | | Crystal structure of alkaline invertase InvA from Anabaena sp. PCC 7120 complexed with glucose | | Descriptor: | Alkaline Invertase, alpha-D-glucopyranose | | Authors: | Xie, J, Cai, K, Hu, H.X, Jiang, Y.L, Yang, F, Hu, P.F, Chen, Y, Zhou, C.Z. | | Deposit date: | 2016-07-28 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural Analysis of the Catalytic Mechanism and Substrate Specificity of Anabaena Alkaline Invertase InvA Reveals a Novel Glucosidase
J. Biol. Chem., 291, 2016
|
|
5GOO
 
 | | Crystal structure of alkaline invertase InvA from Anabaena sp. PCC 7120 complexed with fructose | | Descriptor: | Alkaline Invertase, GLYCEROL, beta-D-fructofuranose | | Authors: | Xie, J, Cai, K, Hu, H.X, Jiang, Y.L, Yang, F, Hu, P.F, Chen, Y, Zhou, C.Z. | | Deposit date: | 2016-07-28 | | Release date: | 2016-11-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structural Analysis of the Catalytic Mechanism and Substrate Specificity of Anabaena Alkaline Invertase InvA Reveals a Novel Glucosidase.
J. Biol. Chem., 291, 2016
|
|
5GVW
 
 | | Crystal structure of the apo-form glycosyltransferase GlyE in Streptococcus pneumoniae TIGR4 | | Descriptor: | Glycosyl transferase family 8, MANGANESE (II) ION | | Authors: | Jiang, Y.L, Jin, H, Zhao, R.L, Yang, H.B, Chen, Y, Zhou, C.Z. | | Deposit date: | 2016-09-07 | | Release date: | 2017-03-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Defining the enzymatic pathway for polymorphic O-glycosylation of the pneumococcal serine-rich repeat protein PsrP.
J. Biol. Chem., 292, 2017
|
|
7VX8
 
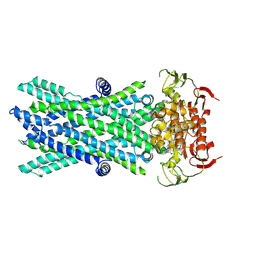 | | Cryo-EM structure of ATP-bound human very long-chain fatty acid ABC transporter ABCD1 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Peroxisomal Membrane Protein related,ATP-binding cassette sub-family D member 1 | | Authors: | Chen, Z.P, Xu, D, Wang, L, Mao, Y.X, Yang, L, Cheng, M.T, Hou, W.T, Chen, Y.X, Zhou, C.Z. | | Deposit date: | 2021-11-12 | | Release date: | 2022-05-18 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis of substrate recognition and translocation by human very long-chain fatty acid transporter ABCD1.
Nat Commun, 13, 2022
|
|
7VWC
 
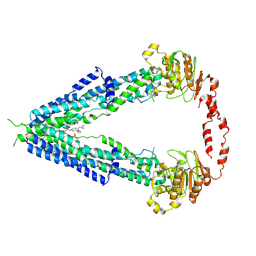 | | Cryo-EM structure of human very long-chain fatty acid ABC transporter ABCD1 | | Descriptor: | Peroxisomal Membrane Protein related,ATP-binding cassette sub-family D member 1, [(2R)-3-[2-azanylethoxy(oxidanyl)phosphoryl]oxy-2-oxidanyl-propyl] octadecanoate | | Authors: | Chen, Z.P, Xu, D, Wang, L, Mao, Y.X, Yang, L, Cheng, M.T, Hou, W.T, Chen, Y.X, Zhou, C.Z. | | Deposit date: | 2021-11-10 | | Release date: | 2022-05-18 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | Structural basis of substrate recognition and translocation by human very long-chain fatty acid transporter ABCD1.
Nat Commun, 13, 2022
|
|
4M00
 
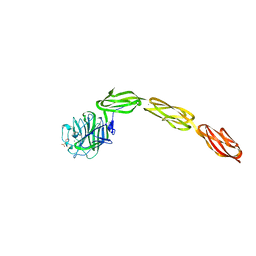 | | Crystal structure of the ligand binding region of staphylococcal adhesion SraP | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, Serine-rich adhesin for platelets, ... | | Authors: | Yang, Y.H, Jiang, Y.L, Zhang, J, Wang, L, Chen, Y, Zhou, C.Z. | | Deposit date: | 2013-08-01 | | Release date: | 2014-06-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural Insights into SraP-Mediated Staphylococcus aureus Adhesion to Host Cells
Plos Pathog., 10, 2014
|
|
4NT9
 
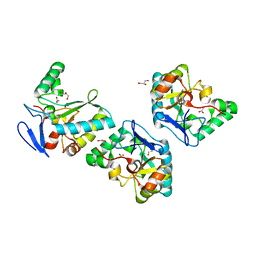 | | Crystal structure of an L,D-carboxypeptidase DacB from Streptococcus pneumonia | | Descriptor: | ACETATE ION, GLYCEROL, Putative uncharacterized protein, ... | | Authors: | Yang, Y.H, Zhang, J, Jiang, Y.L, Zhou, C.Z, Chen, Y. | | Deposit date: | 2013-12-02 | | Release date: | 2014-11-12 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.705 Å) | | Cite: | Crystal structure of an L,D-carboxypeptidase DacB from Streptococcus pneumonia
To be Published
|
|
4OI3
 
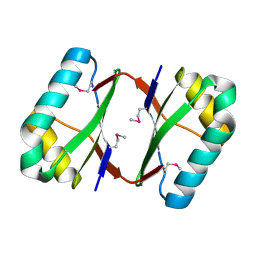 | | Crystal structure analysis of SCO4226 from Streptomyces coelicolor A3(2) | | Descriptor: | Nickel responsive protein | | Authors: | Lu, M, Jiang, Y.L, Wang, S, Cheng, W, Zhang, R.G, Virolle, M.J, Chen, Y, Zhou, C.Z. | | Deposit date: | 2014-01-18 | | Release date: | 2014-09-17 | | Last modified: | 2014-10-22 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Streptomyces coelicolor SCO4226 Is a Nickel Binding Protein.
Plos One, 9, 2014
|
|
4OI6
 
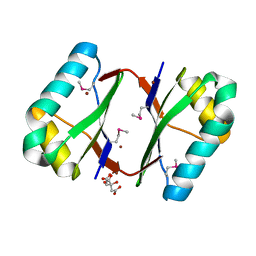 | | Crystal structure analysis of nickel-bound form SCO4226 from Streptomyces coelicolor A3(2) | | Descriptor: | CITRIC ACID, NICKEL (II) ION, Nickel responsive protein | | Authors: | Lu, M, Jiang, Y.L, Wang, S, Cheng, W, Zhang, R.G, Virolle, M.J, Chen, Y, Zhou, C.Z. | | Deposit date: | 2014-01-18 | | Release date: | 2014-09-10 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Streptomyces coelicolor SCO4226 Is a Nickel Binding Protein.
Plos One, 9, 2014
|
|
3M95
 
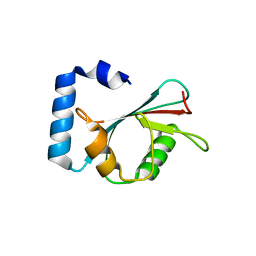 | | Crystal structure of autophagy-related protein Atg8 from the silkworm Bombyx mori | | Descriptor: | Autophagy related protein Atg8 | | Authors: | Teng, Y.-B, Hu, C, Zhang, X, Jiang, Y.L, Hu, H.-X, Zhou, C.Z. | | Deposit date: | 2010-03-20 | | Release date: | 2010-07-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of autophagy-related protein Atg8 from the silkworm Bombyx mori
Acta Crystallogr.,Sect.F, 66, 2010
|
|
4PQG
 
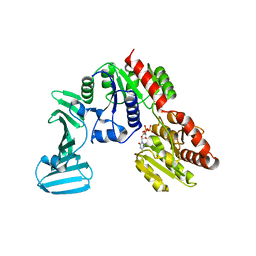 | | Crystal structure of the pneumococcal O-GlcNAc transferase GtfA in complex with UDP and GlcNAc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glycosyltransferase Gtf1, URIDINE-5'-DIPHOSPHATE | | Authors: | Shi, W.W, Jiang, Y.L, Zhu, F, Yang, Y.H, Wu, H, Ren, Y.M, Chen, Y, Zhou, C.Z. | | Deposit date: | 2014-03-03 | | Release date: | 2014-06-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a Novel O-Linked N-Acetyl-d-glucosamine (O-GlcNAc) Transferase, GtfA, Reveals Insights into the Glycosylation of Pneumococcal Serine-rich Repeat Adhesins.
J.Biol.Chem., 289, 2014
|
|
2OE1
 
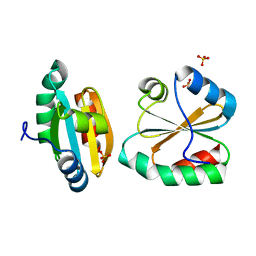 | | Crystal Structure of Mitochondrial Thioredoxin 3 from Saccharomyces cerevisiae (reduced form) | | Descriptor: | SULFATE ION, Thioredoxin-3 | | Authors: | Bao, R, Zhang, Y.R, Zhou, C.Z, Chen, Y.X. | | Deposit date: | 2006-12-28 | | Release date: | 2008-01-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and mechanistic analyses of yeast mitochondrial thioredoxin Trx3 reveal putative function of its additional cysteine residues
Biochim.Biophys.Acta, 1794, 2009
|
|
2OE0
 
 | |
2OE3
 
 | |
4X36
 
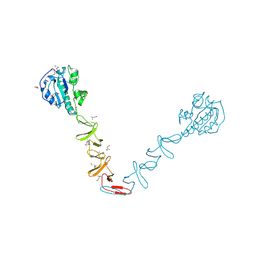 | | Crystal structure of the autolysin LytA from Streptococcus pneumoniae TIGR4 | | Descriptor: | Autolysin, CHOLINE ION, GLYCEROL, ... | | Authors: | Cheng, W, Li, Q, Zhou, C.Z, Chen, Y.X. | | Deposit date: | 2014-11-28 | | Release date: | 2015-05-27 | | Last modified: | 2015-06-24 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Full-length structure of the major autolysin LytA.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
5WXX
 
 | | Crystal structure of Microcystis aeruginosa PCC 7806 aspartate racemase in complex with citrate | | Descriptor: | CITRIC ACID, McyF | | Authors: | Cao, D.D, Zhou, K, Jiang, Y.L, Zhou, C.Z. | | Deposit date: | 2017-01-09 | | Release date: | 2018-01-17 | | Last modified: | 2019-07-31 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural insights into the catalysis and substrate specificity of cyanobacterial aspartate racemase McyF.
Biochem.Biophys.Res.Commun., 514, 2019
|
|
5WXZ
 
 | | Crystal structure of Microcystis aeruginosa PCC 7806 aspartate racemase in complex with D-aspartate | | Descriptor: | D-ASPARTIC ACID, McyF | | Authors: | Cao, D.D, Zhou, K, Jiang, Y.L, Zhou, C.Z. | | Deposit date: | 2017-01-09 | | Release date: | 2018-01-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural insights into the catalysis and substrate specificity of cyanobacterial aspartate racemase McyF.
Biochem.Biophys.Res.Commun., 514, 2019
|
|
4XSP
 
 | | Crystal structure of Anabaena Alr3699/HepE in complex with UDP | | Descriptor: | Alr3699 protein, GLYCEROL, URIDINE-5'-DIPHOSPHATE | | Authors: | Wang, X.P, Dai, Y.N, Jiang, Y.L, Cheng, W, Chen, Y.X, Zhou, C.Z. | | Deposit date: | 2015-01-22 | | Release date: | 2016-01-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural and enzymatic analyses of a glucosyltransferase Alr3699/HepE involved in Anabaena heterocyst envelop polysaccharide biosynthesis
Glycobiology, 26, 2016
|
|
4XSR
 
 | | Crystal structure of Anabaena Alr3699/HepE in complex with UDP-glucose | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Alr3699 protein, SULFATE ION, ... | | Authors: | Wang, X.P, Dai, Y.N, Jiang, Y.L, Cheng, W, Chen, Y.X, Zhou, C.Z. | | Deposit date: | 2015-01-22 | | Release date: | 2016-01-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structural and enzymatic analyses of a glucosyltransferase Alr3699/HepE involved in Anabaena heterocyst envelop polysaccharide biosynthesis
Glycobiology, 26, 2016
|
|
4XSQ
 
 | | Structure of a variable lymphocyte receptor-like protein Bf66946 from Branchiostoma floridae | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, variable lymphocyte receptor-like protein Bf66946 | | Authors: | Cao, D.D, Cheng, W, Jiang, Y.L, Wang, W.J, Li, Q, Chen, Y, Zhou, C.Z. | | Deposit date: | 2015-01-22 | | Release date: | 2016-03-23 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structure of a variable lymphocyte receptor-like protein from the amphioxus Branchiostoma floridae.
Sci Rep, 6, 2016
|
|
